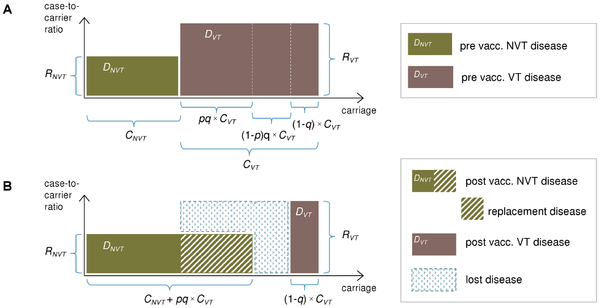
|
|
| Rivi 101: |
Rivi 101: |
|
| |
|
| === [[Työjärjestys]] === | | === [[Työjärjestys]] === |
| [[Työjärjestys|Työjärjestys-sivu]] | | [[Työjärjestys|Työjärjestys-sivulla]] |
|
| |
|
| === Laskenta === | | === [[Mallitus ja laskenta]] === |
| | | [[Työjärjestys|Epidemiologinen malli-sivulla]] |
| R-program code for the replcement model as in File S1 of
| |
| Nurhonen M, Auranen K: Optimal serotype compositions for pneumococcal conjugate vaccination under serotype replacement [[http://www.ploscompbiol.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.1003477]
| |
| | |
| | |
| ==== Esimerkkejä ====
| |
| | |
| * VT: vaccine serotypes, i.e. pneumococcus serotypes that are found in a vaccine.
| |
| * NVT: non-vaccine serotypes, i.e. pneumococcus serotypes that are not found in the vaccine.
| |
| * [http://fi.opasnet.org/fi_wiki/index.php?title=Toiminnot:RTools&id=P6CXptUReneZ8YNh Example model run]
| |
| | |
| {{comment|# |Yritin selvittää kuinka alla oleva koodi toimii. Huomasin että vaihtaessa arvoja ajoon, niin vaikuttaisi siltä että ajon tulokset eivät kuitenkaan muutu? |--[[Käyttäjä:Jaakko|Jaakko]] ([[Keskustelu käyttäjästä:Jaakko|keskustelu]]) 5. kesäkuuta 2014 kello 14.13 (UTC)}}
| |
| {{comment|# |Taulukot vastaavat näköjään aina lähtömallia, joka on PCv13, mutta pylväskuviossa muutettu malli on mukana.| --[[Käyttäjä:Mnud|Markku N.]] ([[Keskustelu käyttäjästä:Mnud|keskustelu]]) 9. kesäkuuta 2014 kello 09.18 (UTC)}}
| |
| | |
| <rcode embed=1 graphics=1 variables="
| |
| name:q_user|description:Proportion of VT carriage removed by vaccination|default:1|
| |
| name:p_user|description:Proportion of VT carriage removed by vaccination that is replaced by NVT|default:1|
| |
| name:adultcarriers|description:The true amount of more than 5-year-old carriers is n times larger than estimated. What is n?|default:1|
| |
| name:servac_user|description:Serotypes in a vaccine to look at|type:checkbox|options:
| |
| '19F';19F;'23F';23F;'6B';6B;'14';14;'9V';9V;'4';4;'18C';18C;'1';1;'7';7;
| |
| '6A';6A;'19A';19A;'3';3;'8';8;'9N';9N;'10';10;'11';11;'12';12;'15';15;
| |
| '16';16;'20';20;'22';22;'23A';23;'33';33;'35';35;'38';38;'6C';6C;'Oth';Other|
| |
| default:'19F';'23F';'6B';'14';'9V';'4';'18C';'1';'7';'6A';'19A';'3'
| |
| ">
| |
| library(OpasnetUtils)
| |
| library(ggplot2)
| |
| | |
| objects.latest("Op_fi4305", code_name = "alusta") # [[Pneumokokkirokote]]
| |
| objects.latest("Op_en6007", code_name = "answer") # [[OpasnetUtils/Drafts]]
| |
| | |
| openv.setN(100)
| |
| | |
| ## Read the annual IPD and carriage incidence data.
| |
| ## The 0 entries in IPD and carriage data are replaced by small values.
| |
| serotypes<-c(
| |
| "19F", "23F", "6B", "14", "9V", "4", "18C", "1", "7",
| |
| "6A", "19A", "3", "8", "9N", "10", "11", "12", "15",
| |
| "16", "20", "22", "23A", "33", "35", "38", "6C", "Oth")
| |
| car_under5<-c(
| |
| 156030, 156030, 126990, 41200, 22290, 12830, 10130, 10, 14180,
| |
| 54940, 24320, 12160, 1350, 20940, 4050, 72270, 10, 33100,
| |
| 3380, 1350, 12160, 3380, 680, 30400, 4050, 27470, 24320 )
| |
| car_over5<-c(
| |
| 168100, 314800, 256700, 209800, 114100, 62500, 200700, 100, 100,
| |
| 158800, 54900, 30800, 8800, 8800, 20800, 97700, 100, 100,
| |
| 191900, 25200, 72500, 22000, 100, 71300, 100, 79400, 330100 )
| |
| ipd_under5<-c(
| |
| 7.78, 7.88, 24.39, 20.76, 2.91, 2.91, 6.64, 0.31, 3.02,
| |
| 3.94, 9.88, 1.25, 0.10, 0.83, 0.41, 0.42, 0.21, 1.98,
| |
| 0.21, 0.01, 0.93, 0.10, 0.42, 0.31, 0.42, 0.01, 0.73 )
| |
| ipd_over5<-c(
| |
| 28.51, 53.72, 29.53, 99.43, 43.07, 76.99, 24.39, 6.58, 46.88,
| |
| 17.42, 20.54, 55.04, 11.21, 25.20, 6.28, 12.76, 13.89, 9.18,
| |
| 4.73, 3.29, 29.03, 4.40, 5.64, 12.41, 1.43, 5.50, 11.20 )
| |
| | |
| ## Combine the data into 2 matrices of dimension 27*2:
| |
| IPD<-cbind(ipd_under5, ipd_over5)
| |
| Car<-cbind(car_under5, car_over5)
| |
| | |
| ## Row numbers corresponding to the 3 different PCV formulations
| |
| ## in matrices IPD and Car. Note: there is no serotype 5 in our data.
| |
| pcv7rows<-seq(7); pcv10rows<-seq(9); pcv13rows<-seq(12)
| |
| | |
| | |
| ## Example S1.2A: Calculate the predicted incidence of IPD for the non-vaccine
| |
| ## types(NVTs) under PCV13. The predictions are calculated separately for the
| |
| ## two age classes. These are the values reported on the bottom panel in
| |
| ## Figure 2 (there given as per 100K incidences).
| |
| postvacc <-Vaccination(IPD,Car,VT_rows=pcv13rows,p=1,q=1)
| |
| | |
| cat("Incidence of invasive pneumococcal disease after vaccination program (p=1, q=1).\n")
| |
| oprint(cbind(serotypes,postvacc[[1]]))
| |
| cat("Number of carriers after vaccination program.\n")
| |
| oprint(cbind(serotypes,postvacc[[2]]))
| |
| | |
| ## Example S1.2B: Decrease in IPD incidence after adding a single new serotype
| |
| ## to PCV13 separately for the two age categories.
| |
| next_under5<-NextVT(IPD[,1],Car[,1], VT_rows=pcv13rows,p=1)
| |
| next_over5 <-NextVT(IPD[,2],Car[,2], VT_rows=pcv13rows,p=1)
| |
| cat("Next serotype to add to a vaccine (p=1).\n")
| |
| oprint(rbind(serotypes, next_under5, next_over5))
| |
| # Nämä taulukot kannattaisi transposata niin näyttäisivät siistimmiltä.
| |
| | |
| ## Example S1.3A: The optimal sequence for under 5 year olds when replacement is 100%.
| |
| ## The output shows the decreases in IPD incidence for each step,
| |
| ## corresponding to Figure 5(C). The last serotype (row 27, the category "Other")
| |
| ## is excluded from any vaccine composition but is taken into account as a
| |
| ## replacing serotype at each stage.
| |
| opt<-OptimalSequence(IPD[,1],Car[,1],VT_rows=0,Excluded_rows=27,p=1.0,HowmanyAdded=20)
| |
| | |
| cat("Optimal sequence of serotypes to add to a vaccine (p=1, HowmanyAdded=20).\n")
| |
| oprint(rbind(serotypes[opt[1,]],opt[2,]))
| |
| | |
| ## Example S1.3B: The optimal sequence for the whole population when
| |
| ## replacement is 50% and the current composition includes the PCV7 serotypes.
| |
| opt<-OptimalSequence(IPD,Car, VT_rows=pcv7rows,Excluded_rows=length(serotypes),
| |
| p=0.5,HowmanyAdded=17)
| |
| | |
| cat("Optimal sequence of serotypes to add to a vaccine (p=0.5, HownamyAdded=17).\n")
| |
| oprint(rbind(serotypes[opt[1,]],opt[2,]))
| |
| | |
| ###################################
| |
| | |
| ## Read the annual IPD and carriage incidence data.
| |
| ## The 0 entries in IPD and carriage data are replaced by small values.
| |
| | |
| IPD <- Ovariable("IPD", ddata = "Op_fi4305.pneumokokki_vaestossa")
| |
| IPD@data <- IPD@data[IPD@data$Observation == "Incidence" , colnames(IPD@data) != "Observation"]
| |
| | |
| Car <- Ovariable("Car", ddata = "Op_fi4305.pneumokokki_vaestossa")
| |
| Car@data <- Car@data[Car@data$Observation == "Carrier" , colnames(Car@data) != "Observation"]
| |
| | |
| servac <- Ovariable("servac", data = data.frame(
| |
| Vaccine = rep(c("Current", "New"), each = length(serotypes)),
| |
| Serotype = serotypes,
| |
| Result = as.numeric(c(
| |
| serotypes %in% c("19F", "23F", "6B", "14", "9V", "4", "18C", "1", "7","6A", "19A", "3"),
| |
| serotypes %in% servac_user
| |
| ))
| |
| ))
| |
| | |
| p <- Ovariable("p", data = data.frame(Result = p_user))
| |
| q <- EvalOutput(Ovariable("q", data = data.frame(Result = q_user)))
| |
| # EvalOutput must be used because q is mentioned twice in the code and there will otherwise be a merge mismatch.
| |
| | |
| # The true number of adult carriers may actually be larger than estimated. This adjusts for that.
| |
| Car <- Car * Ovariable("adjust", data = data.frame(Age = c("Under 5", "Over 5"), Result = c(1, adultcarriers)))
| |
| | |
| VacCar <- EvalOutput(VacCar)
| |
| VacIPD <- EvalOutput(VacIPD)
| |
| | |
| cat("Number of carriers\n")
| |
| oprint(summary(VacCar))
| |
| cat("Incidence of invasive pneumococcal disease.\n")
| |
| oprint(summary(VacIPD))
| |
| | |
| if("Iter" %in% colnames(VacCar@output)) N <- max(VacCar@output$Iter) else N <- 1
| |
| | |
| ggplot(VacCar@output, aes(x = Serotype, weight = result(VacCar) / N, fill = Vaccine)) + geom_bar(position = "dodge") + theme_gray(base_size = 24) +
| |
| labs(title = "Carriers", y = "Number of carriers in Finland")
| |
| | |
| ggplot(VacIPD@output, aes(x = Serotype, weight = result(VacIPD) / N, fill = Vaccine)) + geom_bar(position = "dodge") + theme_gray(base_size = 24) +
| |
| labs(title = "Incidence of invasive pneumococcal disease", y = "Number of cases / 100000 py")
| |
| | |
| ggplot(VacIPD@output, aes(x = Vaccine, weight = result(VacIPD) / N, fill = Age)) + geom_bar(position = "stack") + theme_gray(base_size = 24) +
| |
| labs(title = "Incidence of invasive pneumococcal disease", y = "Number of cases / 100000 py")
| |
| | |
| </rcode>
| |
| | |
| === Funktioiden alustus ===
| |
| | |
| <rcode name="alusta" label="Alusta funktiot" embed=1>
| |
| | |
| library(OpasnetUtils)
| |
| | |
| #S1.4. The R-functions
| |
| ###############################################################################
| |
| ##
| |
| ## R code for the core methods introduced in
| |
| ## Markku Nurhonen and Kari Auranen:
| |
| ## "Optimal serotype compositions for pneumococcal conjugate
| |
| ## vaccination under serotype replacement",
| |
| ## PLoS Computational Biology, 2014.
| |
| ##
| |
| ###############################################################################
| |
| ## List of arguments common to most functions:
| |
| ##
| |
| ## IPD = matrix of IPD incidences by age class (columns) and serotype (rows)
| |
| ## Car = corresponding matrix of carriage incidences
| |
| ## VT_rows = vector of the row numbers in matrices IPD and Car
| |
| ## corresponding to vaccine types (VT_rows=0 for no vaccination)
| |
| ## p = proportion of lost VT carriage which is replaced by NVT carriage
| |
| ## q = proportion of VT carriage lost either due to elimination or replacement
| |
| ##
| |
| ## This code includes 4 functions:
| |
| ## Vaccination, NextVT, OptimalSequence and OptimalVacc.
| |
| ##
| |
| | |
| Vaccination<-function(IPD,Car,VT_rows,p,q) {
| |
| ##
| |
| ## Result:
| |
| ## A list of 2 matrices: IPD and carriage incidences
| |
| ## after vaccination (corresponding to matrices IPD and Car).
| |
| ## [Markku Nurhonen 2013]
| |
| ##
| |
| if (VT_rows[1]>0) {
| |
| IPD<-as.matrix(IPD); Car<-as.matrix(Car)
| |
| # Post vaccination carriage incidences
| |
| Car_Total<-t(matrix(apply(Car,2,sum),dim(Car)[2],dim(Car)[1]))
| |
| Car2<-Car; Car2[VT_rows,]<-0
| |
| Car_NVT<-t(matrix(apply(Car2,2,sum),dim(Car2)[2],dim(Car2)[1]))
| |
| Car_VT<-Car_Total-Car_NVT
| |
| CarNew<-q*(1+p*Car_VT/Car_NVT)*Car2+(1-q)*Car
| |
| # Post vaccination IPD incidences
| |
| NVT_rows<-seq(dim(IPD)[1])[-1*VT_rows]
| |
| # CCR=Case-to-carrier ratios
| |
| CCR<-IPD/Car ; IPDNew<-0*IPD
| |
| # Apply the equation appearing above
| |
| # equation (1) in text for each serotype.
| |
| # First term applies to NVTs.
| |
| IPDNew[VT_rows,]<-(1-q)*IPD[VT_rows,]
| |
| # Second term applies to NVTs.
| |
| IPDNew[NVT_rows,]<-((Car_NVT+p*q*Car_VT)*(Car/Car_NVT)*CCR)[NVT_rows,]
| |
| }
| |
| else {
| |
| IPDNew<-IPD; CarNew<-Car
| |
| }
| |
| list(IPDNew,CarNew)
| |
| }
| |
| | |
| NextVT<-function(IPD,Car,VT_rows,p) {
| |
| ##
| |
| ## Result:
| |
| ## A vector of decreases in IPD due to adding a serotype
| |
| ## to the vaccine. If VT_rows=0, initially no vaccination.
| |
| ## For row indexes incuded in VT_rows, the result is 0.
| |
| ## [Markku Nurhonen 2013]
| |
| ##
| |
| IPD<-as.matrix(IPD); Car<-as.matrix(Car)
| |
|
| |
| ## VaccMat = IPD and Car matrices after vaccination
| |
| VaccMat<-Vaccination(IPD,Car,VT_rows,p,1)
| |
| IPD<-VaccMat[[1]]; Car<-VaccMat[[2]]
| |
|
| |
| ## Total_IPD,Total_Car = Matrices corresponding to
| |
| ## overall IPD and carriage in each age class.
| |
| Total_IPD<-t(matrix(apply(IPD,2,sum),dim(IPD)[2],dim(IPD)[1]))
| |
| Total_Car<-t(matrix(apply(Car,2,sum),dim(Car)[2],dim(Car)[1]))
| |
|
| |
| ## Effect = decrease in IPD when one serotype is added to the vaccine.
| |
| ## See equation (3) in text.
| |
| Effect<-(Total_IPD-IPD)*((IPD/(Total_IPD-IPD))-(p*Car/(Total_Car-Car)))
| |
|
| |
| ## Special case when only one NVT remains.
| |
| IPD_nonzero<-which(apply(IPD,1,sum)!=0)
| |
| if (length(IPD_nonzero)==1) {Effect[IPD_nonzero,]<-IPD[IPD_nonzero,]}
| |
| | |
| ## Result is obtained after summation over age classes.
| |
| apply(Effect,1,sum)
| |
| }
| |
| | |
| OptimalSequence<-function(IPD,Car,VT_rows,Excluded_rows,p,HowmanyAdded) {
| |
| ##
| |
| ## Starting from VTs indicated by the vector VT_rows
| |
| ## (VT_rows=0, for no vaccination) sequentially add new VTs
| |
| ## to the vaccine composition s.t. at each step the optimal
| |
| ## serotype (corresponding to largest decrease in IPD) is added.
| |
| ##
| |
| ## Excluded_rows = Vector of indexes of the rows in matrices
| |
| ## IPD and Car corresponding to serotypes that are not to
| |
| ## be included in a vaccine composition, e.g. a row
| |
| ## corresponding to a group of serotypes labelled "Other".
| |
| ## Enter Excluded_rows=0 for no excluded serotypes.
| |
| ## HowmanyAdded = number of VTs to be added.
| |
| ##
| |
| ## Result:
| |
| ## Matrix of dimension 2*HowmanyAdded with 1st row indicating
| |
| ## the row numbers of added serotypes in the order they appear
| |
| ## in the sequence. The 2nd row lists the decreases in IPD
| |
| ## due to addition of each type. [Markku Nurhonen 2013]
| |
| ##
| |
| IPD<-as.matrix(IPD); Car<-as.matrix(Car)
| |
| ## First check the maximum possible number of added VTs.
| |
| VT_howmany<-length(VT_rows)
| |
| if (VT_rows[1]==0) {VT_howmany<-0}
| |
| Excluded_howmany<-length(Excluded_rows)
| |
| if (Excluded_rows[1]==0) {Excluded_howmany<-0}
| |
| HowmanyAdded<-min(HowmanyAdded,dim(IPD)[1]-(VT_howmany+Excluded_howmany))
| |
| BestVTs<-BestEffects<-rep(0,HowmanyAdded)
| |
| ## Sequential procedure: at each step find the best additional VT.
| |
| for (i in 1:HowmanyAdded) {
| |
| ## Effects = Decrease in IPD after addition of each serotype
| |
| Effects<-NextVT(IPD,Car,VT_rows,p)
| |
| ## Set Effects for VTs and excluded types equal to small values
| |
| ## so that none of these will be selected as the next VT.
| |
| minvalue<- -2*max(abs(Effects))
| |
| if (Excluded_howmany>0) {Effects[Excluded_rows]<-minvalue}
| |
| if (VT_rows[1]>0) {Effects[VT_rows]<-minvalue}
| |
| ## BestVTs[i] = Index of serotype with maximum decrease in IPD.
| |
| BestVTs[i]<-order(-1*Effects)[1]
| |
| ## BestEffects[i] = Decrese in IPD due to addition of BestVTs[i]
| |
| ## to the vaccine.
| |
| BestEffects[i]<-Effects[BestVTs[i]]
| |
| VT_rows<-c(VT_rows,BestVTs[i])
| |
| if (VT_rows[1]==0) {VT_rows<-VT_rows[-1]}
| |
| VaccMat<-Vaccination(IPD,Car,VT_rows,p,1)
| |
| IPD<-VaccMat[[1]]; Car<-VaccMat[[2]]
| |
| }
| |
| t(matrix(c(BestVTs,BestEffects),HowmanyAdded,2))
| |
| }
| |
| | |
| OptimalVacc<-function(IPD,Car,VT_rows,p,q,HowmanyAdded) {
| |
| ##
| |
| ## Result:
| |
| ## A list of 3 elements: (1) Row numbers of serotypes in the optimal
| |
| ## vaccine composition (2)-(3) IPD and carriage incidences
| |
| ## by serotype and age class corresponding to the optimal
| |
| ## vaccine formed using the sequential procedure in the
| |
| ## function OptimalSequence. [Markku Nurhonen 2013]
| |
| ##
| |
| Additional_VTs<-OptimalSequence(IPD,Car,VT_rows,p,HowmanyAdded)[1,]
| |
| All_VTs<-c(VT_rows,Additional_VTs)
| |
| if (All_VTs[1]==0) All_VTs<-All_VTs[-1]
| |
| VaccMat<-Vaccination(IPD,Car,All_VTs,p,q)
| |
| list(All_VTs,VaccMat[[1]],VaccMat[[2]])
| |
| }
| |
| | |
| VacCar <- Ovariable("VacCar",
| |
| dependencies = data.frame(Name = c(
| |
| "IPD", # incidence of pneumococcus disease
| |
| "Car", # number of carriers of pneumococcus
| |
| "servac", # ovariable of serotypes in vaccine (1 for serotypes in a vaccine, otherwise result is 0)
| |
| "p", # proportion of eliminated VT carriage that is replaced by NVT carriage
| |
| "q" # proportion of of VT carriage eliminated by vaccine
| |
| )),
| |
| formula = function(...) {
| |
| ## Result:
| |
| ## An ovariable of carriage incidences
| |
| ## after vaccination (corresponding to Car).
| |
| ## [Markku Nurhonen 2013, Jouni Tuomisto 2014]
| |
| # Post vaccination carriage incidences
| |
|
| |
| # Sum over serotypes and drop extra columns
| |
| Car_Total<- unkeep(oapply(Car, cols = "Serotype", FUN = sum) * 1, prevresults = TRUE)
| |
| # Car2 is a temporary ovariable with NVT carriers only
| |
| Car2 <- unkeep(Car * (1 - servac), prevresults = TRUE) # Take only NVT carriers
| |
|
| |
| Car_NVT <- oapply(Car2, cols = "Serotype", FUN = sum) # Carriers of serotypes not in vaccine (NVT)
| |
| Car_VT <- Car_Total - Car_NVT # Carriers of vaccine serotypes
| |
| | |
| CarNew <- q * (1 + p * Car_VT / Car_NVT) * Car2 + (1 - q) * Car
| |
| | |
| return(CarNew)
| |
| }
| |
| )
| |
|
| |
| VacIPD <- Ovariable("VacIPD",
| |
| dependencies = data.frame(Name = c(
| |
| "IPD", # incidence of pneumococcus disease
| |
| "Car", # number of carriers of pneumococcus
| |
| "servac", # ovariable of serotypes in vaccine (1 for serotypes in a vaccine, otherwise result is 0)
| |
| "p", # proportion of eliminated VT carriage that is replaced by NVT carriage
| |
| "q" # proportion of of VT carriage eliminated by vaccine
| |
| )),
| |
| formula = function(...) {
| |
| ## Result:
| |
| ## An ovariable of IPD incidence
| |
| ## after vaccination (corresponding to ovariable IPD).
| |
| ## [Markku Nurhonen 2013, Jouni Tuomisto 2014]
| |
|
| |
| # Post vaccination carriage incidences (same code as in VacCar)
| |
|
| |
| Car_Total <- unkeep(oapply(Car, cols = "Serotype", FUN = sum) * 1, prevresults = TRUE) # Sums over serotypes
| |
| Car2 <- unkeep(Car * (1 - servac), prevresults = TRUE)
| |
|
| |
| Car_NVT <- oapply(Car2, cols = "Serotype", FUN = sum) # Carriers of serotypes not in vaccine (NVT)
| |
| Car_VT <- Car_Total - Car_NVT # Carriers of vaccine serotypes
| |
| CarNew <- q * (1 + p * Car_VT / Car_NVT) * Car2 + (1 - q) * Car
| |
|
| |
| # Post vaccination IPD incidences
| |
| # CCR=Case-to-carrier ratios
| |
| CCR <- IPD / Car
| |
| | |
| # Apply the equation appearing above
| |
| # equation (1) in text for each serotype.
| |
| # First term applies to VTs.
| |
| IPDNewVT <- (1 - q) * IPD * servac
| |
| | |
| # Second term applies to NVTs.
| |
| IPDNewNVT <- (Car_NVT + p * q * Car_VT) * (Car / Car_NVT) * CCR * (1 - servac)
| |
| | |
| IPDNew <- IPDNewVT + IPDNewNVT
| |
| | |
| return(IPDNew)
| |
| }
| |
| )
| |
| | |
| objects.store(Vaccination, NextVT, OptimalSequence, OptimalVacc, VacCar, VacIPD)
| |
| | |
| cat("Funktiot Vaccination, NextVT, OptimalSequence, OptimalVacc sekä ovariablet VacCar, VacIPD tallennettu. \n")
| |
| | |
| </rcode>
| |
| | |
| === Data ===
| |
| | |
| Pneumokokin esiintyvyys suomalaisessa väestössä. Carrier: (oireettomien) kantajien lukumäärä, Incidence: invasiivisen pneumokokkitaudin ilmaantuvuus 100000 henkilövuotta kohti.
| |
| | |
| <t2b name='Pneumokokki väestössä' index='Serotype,Age,Observation' locations='Carrier,Incidence' unit='#, #/100000py'>
| |
| 19F|Under 5|156030|7.78
| |
| 23F|Under 5|156030|7.88
| |
| 6B|Under 5|126990|24.39
| |
| 14|Under 5|41200|20.76
| |
| 9V|Under 5|22290|2.91
| |
| 4|Under 5|12830|2.91
| |
| 18C|Under 5|10130|6.64
| |
| 1|Under 5|10|0.31
| |
| 7|Under 5|14180|3.02
| |
| 6A|Under 5|54940|3.94
| |
| 19A|Under 5|24320|9.88
| |
| 3|Under 5|12160|1.25
| |
| 8|Under 5|1350|0.1
| |
| 9N|Under 5|20940|0.83
| |
| 10|Under 5|4050|0.41
| |
| 11|Under 5|72270|0.42
| |
| 12|Under 5|10|0.21
| |
| 15|Under 5|33100|1.98
| |
| 16|Under 5|3380|0.21
| |
| 20|Under 5|1350|0.01
| |
| 22|Under 5|12160|0.93
| |
| 23A|Under 5|3380|0.1
| |
| 33|Under 5|680|0.42
| |
| 35|Under 5|30400|0.31
| |
| 38|Under 5|4050|0.42
| |
| 6C|Under 5|27470|0.01
| |
| Oth|Under 5|24320|0.73
| |
| 19F|Over 5|168100|28.51
| |
| 23F|Over 5|314800|53.72
| |
| 6B|Over 5|256700|29.53
| |
| 14|Over 5|209800|99.43
| |
| 9V|Over 5|114100|43.07
| |
| 4|Over 5|62500|76.99
| |
| 18C|Over 5|200700|24.39
| |
| 1|Over 5|100|6.58
| |
| 7|Over 5|100|46.88
| |
| 6A|Over 5|158800|17.42
| |
| 19A|Over 5|54900|20.54
| |
| 3|Over 5|30800|55.04
| |
| 8|Over 5|8800|11.21
| |
| 9N|Over 5|8800|25.2
| |
| 10|Over 5|20800|6.28
| |
| 11|Over 5|97700|12.76
| |
| 12|Over 5|100|13.89
| |
| 15|Over 5|100|9.18
| |
| 16|Over 5|191900|4.73
| |
| 20|Over 5|25200|3.29
| |
| 22|Over 5|72500|29.03
| |
| 23A|Over 5|22000|4.4
| |
| 33|Over 5|100|5.64
| |
| 35|Over 5|71300|12.41
| |
| 38|Over 5|100|1.43
| |
| 6C|Over 5|79400|5.5
| |
| Oth|Over 5|330100|11.2
| |
| </t2b>
| |
|
| |
|
| == Katso myös == | | == Katso myös == |