|
|
| Rivi 1: |
Rivi 1: |
| [[Luokka:Projekti]] | | [[Luokka:Projekti]] |
| {{arviointi|moderaattori=Jouni}} | | {{arviointi|moderaattori=Jouni}} |
| '''Ruori''' on VN-TEAS-hanke, jossa arvioidaan erilaisia ruokaan liittyviä riskitekijöitä, niiden vähentämispotentiaalia ja niiden terveys- ja talousvaikutuksia. | | '''Ruori''' on VN-TEAS-hanke, jossa arvioidaan erilaisia ruokaan liittyviä riskitekijöitä, niiden vähentämispotentiaalia ja niiden terveys- ja talousvaikutuksia. Hankkeen [http://urn.fi/URN:ISBN:978-952-287-796-3 loppuraportti] on julkaistu 2019. |
|
| |
|
| == Rajaus == | | == Rajaus == |
| Rivi 50: |
Rivi 50: |
| Toinen tapa on tehdä kokeluja toisaalta tuotantoprosessissa (kalarehuvalmisteiden puhdistamista dioksiineista kokeiltiin aikanaan menestyksekkäästi, joten se on Suomessa nykyään tavallista eikä pitoisuuksien valvonta ei ole kovin tarpeellista) ja toisaalta tehokkaimman valvontapisteen tunnistamisessa (HACCP eli hazard analysis and critical control point on edelleen hyvä käytäntö). | | Toinen tapa on tehdä kokeluja toisaalta tuotantoprosessissa (kalarehuvalmisteiden puhdistamista dioksiineista kokeiltiin aikanaan menestyksekkäästi, joten se on Suomessa nykyään tavallista eikä pitoisuuksien valvonta ei ole kovin tarpeellista) ja toisaalta tehokkaimman valvontapisteen tunnistamisessa (HACCP eli hazard analysis and critical control point on edelleen hyvä käytäntö). |
|
| |
|
| Ruokavalion osalta tilanne on erilainen. Ei ole pelkoa, että jokin ruoka yllättäen sisältäisi enemmän suolaa tai vähemmän hedelmää kuin toisen erän valmiste; terveyteen vaikuttavat tekijät ovat siis hyvin ennakoitavissa ja vakioitu. Haasteet ovat yksilöiden käyttäytymisen ohjailussa niin, että hänen kokonaisruokavalionsa olisi terveellinen. Yksittäisen tuotteen suosiminen tai rajoittaminen ei riitä. Vaikuttamisen keinot kuten verotus myös ovat karkeita eli eivät kohdistu tehokkaasti oikeisiin yksilöihin tai asioihin. | | Ruokavalion osalta tilanne on erilainen. Ei ole pelkoa, että jokin ruoka yllättäen sisältäisi enemmän suolaa tai vähemmän hede |
| | |
| Tässäkin kokeilut voisivat tuottaa uudenlaista lisäarvoa. Kansalaispalkkakokeilu toi suppeudestaan huolimatta arvokasta tietoa siitä, mihin kansalaispalkka vaikuttaa ja mihin ei. Samalla tavalla terveellisen ruoan verokohtelua, sokeriveroa tai sydänmerkkiaterioita ja niiden tehoa ja toimivuutta pitäisi yrittää kokeilujen avulla mitata ja onnistuneita keinoja ottaa laajaan käyttöön. Elintarvike- ja kaupan ala toki tekee jatkuvasti omia kokeilujaan ja lienee varsin hyvin perillä siitä, miten joidenkin tuotteiden myyntiä voidaan lisätä sopivalla sijoittelulla, hinnoittelulla ja pakkauksilla. Tässä tavoitteet vain ovat kansanterveyden kannalta osittain ristiriitaiset. On myös vaikea keksiä, miten nämä liikesalaisuudet voitaisiin saada yhteiskunnan käyttöön terveyden edistämiseksi.
| |
| | |
| Kun tarkastellaan eri maiden tilannetta ja ruoan ravitsemuksellisia ja hygieenisiä riskejä kokonaisuutena, voidaan tehdä kokoava päätelmä. Valvonta on hyödyllisimmillään eli tieto arvokkaimmillaan silloin, kun toiminnan, raaka-aineiden tai tuotteiden laatu vaihtelee paljon. Silloin valvonnan avulla voidaan tehokkaasti ohjata toimintaa turvallisemmaksi esimerkiksi hylkäämällä saastuneita eriä. Huonon hygienian oloissa tiedetään käsien pesun välttämättömyys ilman mittauksiakin, ja huippuunsa viritetyssä elintarvikeketjussa syntyy kovin vähän hylättävää. Ravitsemuspuolella laadun vaihtelu ei ole elintarvikkeissa vaan ihmisten tavassa käyttää epäterveellisiä tuotteita. Myös ravitsemusriskien kokemisessa ja tietämyksessä lienee erittäin suuria yksilöllisiä eroja. Näiden tutkiminen ja ymmärtäminen auttaisi suunnittelemaan sellaisia kansanterveyttä parantavia toimia, joilla ei kuitenkaan rajoitettaisi liikaa ihmisten mahdollisuuksia syödä myös herkkuja.
| |
| | |
| == Perustelut ==
| |
| | |
| === Data ===
| |
| | |
| Mitä kaikkea kuuluu vähäiseen hedelmien tai vihannesten syöntiin?
| |
| * Vähähedelmäinen ruokavalio: hedelmien kulutus alle 3 annosta päivässä (310 g yhteensä) (sisältää tuoreet, pakastetut, keitetyt, säilötyt ja kuivatut hedelmät mutta ei sisällä hedelmämehuja tai suolaan tai etikkaan säilöttyjä hedelmiä) http://www.healthdata.org/terms-defined. Diet low in fruits: Consumption of less than 3 servings (310 g total) of fruits per day (includes fresh, frozen, cooked, canned, or dried fruit but excludes fruit juices and salted or pickled fruits).
| |
| * Vähävihanneksinen ruokavalio: vihannesten kulutus alle 4 annosta (400 g yhteensä) (sisältää tuoreet, pakastetut, keitetyt, säilötyt ja kuivatut vihannekset mukaan lukien palkokasvit mutta ei sisällä suolaan tai etikkaan säilöttyjä vihanneksia eikä pähkinöitä, siemeniä tai tärkkelyspitoisia vihanneksia kuten perunaa tai maissia). Diet low in vegetables: Consumption of less than 4 servings (400 g total) of vegetables per day (includes fresh, frozen, cooked, canned, or dried vegetables including legumes but excluding salted or pickled, juices, nuts and seeds, and starchy vegetables such as potatoes or corn).
| |
| | |
| Luken tilastoista löytyy tietoja kalansyönnistä Suomessa. Järvikalaa ei ole eritelty, mutta muut kuin viljellyt ja merilajit ovat yhteensä 2.6 kg/a henkeä kohti. [https://stat.luke.fi/en/fish-consumption-2017_en]. Kalansyöntidatat löytyvät myös Opasnetistä [[:op_en:Goherr: Fish consumption study]].
| |
| | |
| Voiko DALYt muuntaa euroiksi, ja miten se tehdään?
| |
| * Drake ehdottaa globaalin arvon päättämistä DALYn hinnaksi, samaan tapaan kuin 1,25 dollarin alittava päivätulo on määritelty absoluuttiseksi köyhyydeksi. Tällöin kaikki tuota hintaa kustannustehokkaammat toimet kannattaisi tehdä joko kansallisin, tai jos se ei jostain syystä onnistu, kansainvälisin toimin. Hän ei kuitenkaan ehdota suuruutta tälle hinnalle.<ref>Drake T. (2014) Priority setting in global health: towards a minimum DALY value. Health Economics Letter 23:2:248-252. https://doi.org/10.1002/hec.2925</ref>
| |
| * Brent on analysoinut implisiittisiä hintoja DALYlle Global Fund for AIDS, Tuberculosis, and Malaria -säätiön rahoituspäätöksistä. DALYn hinta näyttää olevan 6300 USD kaikille taudeille keskimäärin, ja 11900 USD erityisesti HIV/AIDSille<ref>Brent RJ. (2011) An implicit price of a DALY for use in a cost-benefit analysis of ARVs. Applied Economics 43:11:1413-1421. https://doi.org/10.1080/00036840802600475</ref>. Globaalit luvut ovat toki pienemmät kuin mitä rikkaissa länsimaissa katsottaisiin aiheelliseksi käyttää.
| |
| * Erilaisista arvioinneista löytyy vaihtelevia lukuja yhden DALYn rahalliselle hinnalle. Esimerkiksi IOMin Shecan-projekti käytti arvoa 50393 €/menetetty elinvuosi<ref>Minstry R. (2011) Methodology for valuing health impacts on the SHEcan project. IOM Research Project P937/96. http://ec.europa.eu/social/BlobServlet?docId=10178&langId=en</ref>, ja IGCB(N)-meluarviointiryhmä käyttää arvoa 60000 GBP/QALY (laatupainotettu elinvuosi) mutta samalla toteaa, että eri arvioinneissa arvot voivat vaihdella välillä 29000 - 130000 GBP/QALY<ref>The Interdepartmental Group on Costs and Benefits Noise Subject Group. (2014) Environmental Noise: Valuing impacts on sleep disturbance, annoyance, hypertension, productivity and quiet. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/380852/environmental-noise-valuing-imapcts-PB14227.pdf</ref><ref>The Interdepartmental Group on Costs and Benefits Noise Subject Group (IGCB(N)). (2010) Noise & Health – Valuing the Human Health Impacts of Environmental Noise Exposure. https://khub.net/c/document_library/get_file?uuid=6a229977-e27a-43c5-a780-e224649bd2df&groupId=6197021</ref>.
| |
| * Berryn ja Flindellin mukaan Isossa-Britanniassa käytäntö on muodostunut sellaiseksi, että lääkkeet tai muut lääketieteelliset toimenpiteet saavat kansallisessa terveysjärjestelmässä helposti puollon, jos ne tuottavat yhden terveen elinvuoden alle 20000 GBP:n kustannuksilla. Tyypillisesti toteutetaan hankkeita tasolla 30000 GBP/QALY, mutta hankkeilta hinnaltaan yli 50000 GBP/QALY vaaditaan erityisiä perusteluja<ref>Berry BF, Flindell IH. (2009) Estimating Dose-Response Relationships between Noise Exposure and Human Health Impacts in the UK. BEL Technical Report 2009-002. https://webarchive.nationalarchives.gov.uk/20130123222353/http://archive.defra.gov.uk/environment/quality/noise/igcb/documents/tech-report.pdf</ref>.
| |
| * Toisaalta Hammitt todistelee, että hinta per tilastollinen elämä (value per statistical life, VSL) ja hinta per DALY muuttuvat epälineaarisesti suhteessa toisiinsa, eikä näin ollen olisi mahdollista käyttää hyvinvointimuutoksen mittarina vakiolla kerrottua DALY-arvoa, ainakaan taloudellisen hyvinvointiteorian (economic welfare theory) puitteissa.<ref>Hammitt, J.K. (2013) Admissible utility functions for health, longevity, and wealth: integrating monetary and life-year measures. J Risk Uncertain 47: 311. https://doi.org/10.1007/s11166-013-9178-4</ref>
| |
| | |
| Pitoisuusanalyysien kustannukset on poistettu tästä.
| |
| | |
| Trikiinin valvontakustannukset: Tämän artikkelin mukaan trikiinin DALYt ovat vain luokkaa 100 DALY/miljardi ihmistä, joten valvonta ei ole mielekästä<ref>Brecht Devleesschauwer, Nicolas Praet, Niko Speybroeck, Paul R. Torgerson, Juanita A. Haagsma, KrisDe Smet, K. Darwin Murrell, Edoardo Pozio, Pierre Dorny. (2015) The low global burden of trichinellosis: evidence and implications. International Journal for Parasitology 45, 2–3, 95-99. [https://doi.org/10.1016/j.ijpara.2014.05.006] [https://www.sciencedirect.com/science/article/pii/S0020751914001374]</ref>.
| |
| | |
| ==== Skenaarioista ====
| |
| | |
| Tyydyttyneen rasvan ja suolan osalta tarkastellaan sydänmerkkiaterioita. Oletetaan, että kaikki lounasruokaloiden ateriat muuttuvat sydänmerkkiaterioiksi. Väestön suuruudeksi oletetaan nykyinen lounaspaikkaruokailijoiden päivittäinen määrä eli 279000, ja heille oletetaan pysyvä muutos ruokavalioon.
| |
| | |
| Kun ei murehdita altistusjakaumasta, voidaan keskimääräisestä saannista vähentää vähennys, ja tämä muutos kohdennetaan vain 25-69-vuotiaaseen alaryhmään (jos alaryhmä on kyseiselle vasteelle määritelty). Skenaariossa kerrotaan PAF luvulla, joka saa arvon 1 nykyisellä altistuksella ja arvon 0 suosituksen mukaisella arvolla. Tätäitä laimennetaan altistujien osuudella. Niinpä käytetään tätä kaavaa kertoimen laskemiseksi:
| |
| PAF_factor = 1-(reduction / (intake-recommendation)) * eaters / population
| |
| | |
| '''Suola:
| |
| | |
| Sydänmerkki vähensi 2016 suolan saantia 1 g/d eli 4.2-5.2 g/vk eli 10 %. Tässä oletetaan 3.3-5.1 g/vk vähennys eli 0.471 - 0.729 g/d myös viikoloput huomioiden. Jula (2011) arvioi, että 1 g/d vähennys koko väestössä alentaa kustannuksia 70 Me/a. (Tarkista onko tautitaakkaa!)
| |
| | |
| Suositus on enintään 5 g/d. Nykysaanti (Valsta 2018) on miehillä 8.7 g/d ja naisilla 6.4 g/d ja tätä käytetään tasajakaumana koko väestölle, koska sukupuolia ei tarkastella erikseen.
| |
| | |
| '''Rasva:
| |
| | |
| Sydänmerkkiateria vähensi saantia 14 E%:sta 10 E%:iin. Tässä oletetaan, että vähennys on 22.0-58.1 g/vk.
| |
| | |
| Suositus on enintään 10 E%.
| |
| | |
| Valsta (2018): Tyydyttyneiden rasvahappojen osuus kokonaisenergiasta oli naisilla 14 % (28 g/vrk) ja miehillä 15 % (38 g/vrk). Naisilla muuntosuhde on 2 g /d /E% ja miehillä 2.53 g /d /E%, käytetään 2.25 g /d /E%. Grammamääräinen altistus on muutettava energiaosuudeksi.
| |
| 22.0-58.1 g/vk / (7d/vk) / (2.25 g /d /E%) = 1.397 - 3.689 E%
| |
| | |
| Näiden tietojen avulla lasketaan PAF-kertoimen jakauma, joka sijoitetaan päätöstauluun päätöksenä Scenario ja vaihtoehtona Action.
| |
| | |
| ==== Malliparametrit ====
| |
| | |
| Malliparametrit saat näkyviin klikkaamalla.
| |
| | |
| {{piilotettu|
| |
| | |
| <t2b name="Malliparametrit" index="Response,Exposure_agent,Type,Subgroup,Unit" obs="Result" desc="Description" unit="various">
| |
| CHD death||BoD|Age:Female 25-69|DALY|9876 (9103 - 10784)|Z:\Projects\RUORI\tautitaakka\Rasvat\IHD_data_IHME.csv
| |
| CHD death||BoD|Age:Male 25-69|DALY|48851 (54035 - 46123)|Z:\Projects\RUORI\tautitaakka\Rasvat\IHD_data_IHME.csv
| |
| CHD death||BoD|Age:Age 25-69|DALY|58727 (63138 - 56907)|Summed from previous
| |
| CHD death||BoD|Age:Female 70+|DALY|42750 (41007 - 45909)|Z:\Projects\RUORI\tautitaakka\Rasvat\IHD_data_IHME.csv
| |
| CHD death||BoD|Age:Male 70+|DALY|48150 (46327 - 51255)|Z:\Projects\RUORI\tautitaakka\Rasvat\IHD_data_IHME.csv
| |
| CHD death||BoD|Age:Age 70+|DALY|90900 (87334 - 97164)|Summed from previous
| |
| Diet high in sodium||BoD|Age:Total population|DALY|1310 : 27670 : 66420|IHME GBD2017. Triangular distribution used because normal would go below zero
| |
| Diet low in fruits||BoD|Age:Total population|DALY|36050 (20570 - 54800)|IHME GBD2017 http://ghdx.healthdata.org/gbd-results-tool?params=gbd-api-2017-permalink/da3bde44e863adb438c5fb47a89942fb
| |
| Diet low in vegetables||BoD|Age:Total population|DALY|28440 (14190 - 45960)|IHME GBD2017
| |
| Liver cancer||BoD|Age:Age 25-64|DALY|2499 (2018 - 3115)|From IHME (2014)
| |
| Liver cancer||BoD|Age:Age 65-74|DALY|2745 (2245 - 3311)|From IHME (2014)
| |
| dummy||case burden|Age:Female 25-69|DALY /case|0|Needed for case_burden to cover all Ages
| |
| dummy||case burden|Age:Female 70+|DALY /case|0|Needed for case_burden to cover all Ages
| |
| dummy||case burden|Age:Male 25-69|DALY /case|0|Needed for case_burden to cover all Ages
| |
| dummy||case burden|Age:Male 70+|DALY /case|0|Needed for case_burden to cover all Ages
| |
| dummy||case burden|Age:Age 25-64|DALY /case|0|Needed for case_burden to cover all Ages
| |
| dummy||case burden|Age:Age 65-74|DALY /case|0|Needed for case_burden to cover all Ages
| |
| IQ loss||case burden|Age:Age 1|DALY /IQ|0.11 (0.06 - 0.16)|Arja used 0.013 but here we use Goherr value instead
| |
| Listeriosis||case burden|Age:Total population|DALY/case|10 (5 - 13.3)|WHO 2015 report, European values (Table A8.2)
| |
| Noro infection||case burden|Age:Total population|DALY/case|0.0015 - 0.0025|WHO 2015 report, European values (Table A8.2)
| |
| Toxoplasmosis||case burden|Age:Age 0 (congenital)|DALY/case|7 (3 - 10)|WHO 2015 report, European values (Table A8.4) 2 (1-3) / 0.3
| |
| Toxoplasmosis||case burden|Age:Age 1+ (acquired)|DALY/case|0.05 (0.03 - 0.08)|WHO 2015 report, European values (Table A8.4) 6 (4 - 10) / 119
| |
| Cancer morbidity||case burden|Age:Female 18-45|DALY/case|0 - 0.28|Goherr assessment
| |
| Cancer morbidity||case burden|Age:Non female 18-45|DALY/case|0 - 0.28|Goherr assessment
| |
| Sperm concentration||case burden|Age:Age 1|DALY/case|0 - 5|Goherr assessment
| |
| Yes or no dental defect||case burden|Age:Age 1|DALY/case|0 - 0.12|Goherr assessment
| |
| |Aflatoxin|exposure|Exposure:To eater; Age:Age 25-64|ng /kg /d|0.85 - 1.14|Finravinto 2012
| |
| |Aflatoxin|exposure|Exposure:To eater; Age:Age 65-74|ng /kg /d|0.5 - 0.67|Finravinto 2012
| |
| |TEQ|exposure|Exposure:To child; Age: Age 1|pg /g|1.65 (0.38 - 3.47)|Ruori code; data from Goherr assessment
| |
| |TEQ|exposure|Exposure:To eater; Age: Female 18-45|pg /d|4.79 (1.07 - 11.48)|Ruori code; data from Goherr assessment
| |
| |TEQ|exposure|Exposure:To eater; Age: Non female 18-45|pg /d|22.61 (9.43 - 44.75)|Ruori code; data from Goherr assessment
| |
| |Lead|exposure|Exposure:To eater; Age:Age 1|ug /l|2:12:30|Measured as blood concentration, triangular distribution for Age 1. RASKURI, Z:\Projects\RUORI\tautitaakka\Lyijy\Lyijy_tautitaakkadata.xlsx
| |
| |Saturated fat|exposure|Exposure:To eater; Age:Age 25-69|E%|13.1 (12.9 - 13.4)|Finland, 2010 situation from Wang et al. Supplementary
| |
| |Saturated fat|exposure|Exposure:To eater; Age:Age 70+|E%|13.2 (12.8 - 13.6)|Finland, 2010 situation from Wang et al. Supplementary
| |
| |Aflatoxin|frexposed|Age:Age 25-64|fraction|1|dummy variable
| |
| |Aflatoxin|frexposed|Age:Age 65-74|fraction|1|dummy variable
| |
| |Lead|frexposed|Age:Age 1|fraction|1|Already in the exposure distribution
| |
| |TEQ|frexposed|Age:Age 1|fraction|1|frexposed is already in the exposure distribution
| |
| |TEQ|frexposed|Age:Female 18-45|fraction|1|frexposed is already in the exposure distribution
| |
| |TEQ|frexposed|Age:Non female 18-45|fraction|1|frexposed is already in the exposure distribution
| |
| IQ loss||incidence|Age:Age 1|IQ /100000py|596000|On average, a population has ca. 6 IQ points per person below 100: mean(abs(rnorm(10000, 100,15)-100))/2
| |
| Listeriosis||incidence|Age:Total population|# /100000py|1.22|Tartuntatautirekisteri 2016: 66 kpl. WHO 2015 report, European values (Table A8.2): 0.2 (0.2 - 0.3)
| |
| Liver cancer||incidence|Age:Age 25-64|# /100000py|4.06|Finnish Cancer Registry, average 2011-2015
| |
| Liver cancer||incidence|Age:Age 65-74|# /100000py|26.16|Finnish Cancer Registry, average 2011-2015
| |
| Noro infection||incidence|Age:Total population|# /100000py|1652 (630 - 3294)|WHO 2015 report, European values (Table A8.2)
| |
| Toxoplasmosis||incidence|Age:Age 0 (congenital)|# /100000py|35 (23 - 81)|WHO 2015 report, European values (Table A8.4) Numbers are per whole population, so scale up to Age0: 0.3 (0.2 - 0.7) * 5517919/47663
| |
| Toxoplasmosis||incidence|Age:Age 1+ (acquired)|# /100000py|119 (79 - 188)|WHO 2015 report, European values (Table A8.4)
| |
| Cancer morbidity||incidence|Age:Female 18-45|# /100000py|657|Statistics Finland https://tilastot.syoparekisteri.fi/syovat/ applies to all subgroups because cancer is lifetime risk
| |
| Cancer morbidity||incidence|Age:Non female 18-45|# /100000py|657|Statistics Finland https://tilastot.syoparekisteri.fi/syovat/ applies to all subgroups because cancer is lifetime risk
| |
| Sperm concentration||incidence|Age:Age 1|# /100000py|7000|Male infertility rate is 7 % (Wikipedia)
| |
| Yes or no dental defect||incidence|Age:Age 1|# /100000py|22400|Alaluusua et al 2004 found 11/49 cases in two lowest groups
| |
| Diet low in fruits|Fruits|PAF|Age:Total population|fraction|1|dummy variable
| |
| Listeriosis|Listeria|PAF|Age:Total population|fraction|1|dummy variable
| |
| Noro infection|Noro virus|PAF|Age:Total population|fraction|1|dummy variable
| |
| CHD death|Saturated fat|PAF|Age:Age 25-69|fraction|0.064 (0.050 - 0.078)|Finland, 2010 situation from Wang et al. Supplementary
| |
| CHD death|Saturated fat|PAF|Age:Age 70+|fraction|0.048 (0.033 - 0.063)|Finland, 2010 situation from Wang et al. Supplementary
| |
| Diet high in sodium|Sodium|PAF|Age:Total population|fraction|1|dummy variable
| |
| Toxoplasmosis|Toxoplasma gondii|PAF|Age:Age 0 (congenital)|fraction|1|dummy variable
| |
| Toxoplasmosis|Toxoplasma gondii|PAF|Age:Age 1+ (acquired)|fraction|1|dummy variable
| |
| Diet low in vegetables|Vegetables|PAF|Age:Total population|fraction|1|dummy variable
| |
| ||population|Age:Age 1||50934|Statistics Finland, 2018 https://pxnet2.stat.fi:443/PXWeb/sq/ac3373d0-e303-4c67-b32a-73c6d26df809
| |
| ||population|Age:Age 25-64|#|2814305|Statistics Finland
| |
| ||population|Age:Age 65-74|#|692868|Statistics Finland
| |
| ||population|Age:Age 25-69|#|3176513|Statistics Finland
| |
| ||population|Age:Age 70+|#|842629|Statistics Finland
| |
| ||population|Age:Total population|#|5517919|Statistics Finland
| |
| ||population|Age:Age 0 (congenital)|#|47663|Statistics Finland
| |
| ||population|Age:Age 1+ (acquired)|#|5470256|Statistics Finland
| |
| ||population|Age:Female 18-45|#|923697|Statistics Finland
| |
| ||population|Age:Non female 18-45|#|4594222|Statistics Finland
| |
| Hepatitis||prevalence|Hepatitis:Hepatitis B-|fraction|0.005|TerveSuomi
| |
| Hepatitis||prevalence|Hepatitis:Hepatitis B+|fraction|0.995|TerveSuomi
| |
| |Saturated fat|scenario exposure|Age:Male 18-24|E%|14.9|Finravinto 2017. Supplementary table 7.12. Average daily intake of saturated fats by gender and age.
| |
| |Saturated fat|scenario exposure|Age:Male 25-44|E%|15.2|Finravinto 2017. Supplementary table 7.12. Average daily intake of saturated fats by gender and age.
| |
| |Saturated fat|scenario exposure|Age:Male 45-64|E%|15.3|Finravinto 2017. Supplementary table 7.12. Average daily intake of saturated fats by gender and age.
| |
| |Saturated fat|scenario exposure|Age:Male 65-74|E%|14.4|Finravinto 2017. Supplementary table 7.12. Average daily intake of saturated fats by gender and age.
| |
| |Saturated fat|scenario exposure|Age:Female 18-24|E%|13.8|Finravinto 2017. Supplementary table 7.12. Average daily intake of saturated fats by gender and age.
| |
| |Saturated fat|scenario exposure|Age:Female 25-44|E%|14.8|Finravinto 2017. Supplementary table 7.12. Average daily intake of saturated fats by gender and age.
| |
| |Saturated fat|scenario exposure|Age:Female 45-64|E%|14.3|Finravinto 2017. Supplementary table 7.12. Average daily intake of saturated fats by gender and age.
| |
| |Saturated fat|scenario exposure|Age:Female 65-74|E%|14.0|Finravinto 2017. Supplementary table 7.12. Average daily intake of saturated fats by gender and age.
| |
| IQ loss|Lead|threshold|Age:Age 1|ug /l|0-24|Lanphear et al 2005 https://doi.org/10.1289/ehp.7688 CHECK THRESHOLD
| |
| Liver cancer|Aflatoxin|UR|Hepatitis:Hepatitis B-|# /(ng /kg /d /100000py)|0.01 (0.002 - 0.03)|WHO Is this per year or per lifetime?
| |
| Liver cancer|Aflatoxin|UR|Hepatitis:Hepatitis B+|# /(ng /kg /d /100000py)|0.3 (0.01 - 0.5)|WHO Is this per year or per lifetime?
| |
| IQ loss|Lead|UR|Age:Age 1|IQ l /ug|0.039 (0.024 - 0.053)|Lanphear et al 2005 https://doi.org/10.1289/ehp.7688 using the first increment from 24 to 100 ug/l
| |
| |Sodium|reduction|Age:Total population|g /d|0.471 - 0.729|RUORI estimates 3.3-5.1 g/week reduction for workplace lunches with heart-friendly label
| |
| |Sodium|intake|Age:Total population|g /d|6.4 - 8.7|Valsta 2018 estimated the lower value for females and upper for males.
| |
| |Sodium|recommendation|Age:Total population|g /d|5|Finnish recommendation by Ruokavirasto
| |
| |Sodium|eaters|Age:Total population|#|279000|RUORI estimate based on Taloustutkimus 2010
| |
| |Saturated fat|reduction|Age:Age 25-69|E%|1.397 - 3.689|RUORI estimates 22.0 - 58.1 g /week, and on average 2.25 g /d /E%
| |
| |Saturated fat|intake|Age:Age 25-69|E%|14 - 15|Valsta 2018 estimated the lower value for females and upper for males.
| |
| |Saturated fat|recommendation|Age:Age 25-69|E%|10|Recommendation by Ruokavirasto?
| |
| |Saturated fat|eaters|Age:Age 25-69|#|279000|RUORI estimate based on Taloustutkimus 2010
| |
| |Vegetables|reduction|Age:Total population|g /d|-30 - 0|RUORI estimates that the consumption increases by 0-14% (up to the % change of tax reduction) i.e. 0.14*213.
| |
| |Vegetables|intake|Age:Total population|g /d|196 - 213|Valsta 2018 estimated the lower value for males and upper for females. (vegetables, legumes, nuts)
| |
| |Vegetables|recommendation|Age:Total population|g /d|400|Recommendation by IHME
| |
| |Vegetables|eaters|Age:Total population|#|5470256|RUORI estimate based on Taloustutkimus 2010
| |
| |Fruits|reduction|Age:Total population|g /d|-26 - 0|RUORI estimates that the consumption increases by 0-14% (up to the % change of tax reduction) i.e. 0.14*189.
| |
| |Fruits|intake|Age:Total population|g /d|135 - 189|Valsta 2018 estimated the lower value for males and upper for females. (fruits and berries)
| |
| |Fruits|recommendation|Age:Total population|g /d|310|Recommendation by IHME
| |
| |Fruits|eaters|Age:Total population|#|5470256|RUORI estimate based on Taloustutkimus 2010
| |
| </t2b>
| |
| | |
| Annos-vasteet on tässä vain näytillä, ja oikeat käyttöön tulevat luvut ovat sivulla [[:op_en:ERFs of environmental pollutants]].
| |
| | |
| <t2b name="Decisions" index="Decision,Option,Variable,Cell,Change" obs="Result" desc="Description" unit="-">
| |
| Adjust|BAU|incidence||Multiply|0.00001|1/100000 py --> 1 py
| |
| Adjust|BAU|PAF|Exposure_agent:Saturated fat|Multiply|1.132|Scaled from Wang to Finravinto 2017: mean(15.2,15.3,14.8,14.0)/13.1
| |
| Hepatitis|Hepatitis B-|BoD||Multiply|0.995|Healthy people. Data from TerveSuomi
| |
| Hepatitis|Hepatitis B+|BoD||Multiply|0.005|Hepatitis B patients. Data from TerveSuomi
| |
| Adjust|BAU|ERF|Response:Liver cancer|Multiply|0.00001|1/100000py --> 1/py
| |
| Scenario|Action|PAF|Exposure_agent:Saturated fat|Multiply|0.911 : 0.944 : 0.971|Based on RUORI modelling (see code)
| |
| Scenario|Action|PAF|Exposure_agent:Sodium|Multiply|0.975 : 0.990 : 0.993|Based on RUORI modelling (see code)
| |
| Scenario|BAU|PAF||Identity||For completion
| |
| Scenario|Action|PAF|Response:Toxoplasmosis;Age:Age 0 (congenital)|Multiply|0|Complete testing and treatment of gongenital disease
| |
| Scenario|Action|PAF|Response:Listeriosis|Multiply|0|Complete testing and removal of pathogen
| |
| Scenario|Action|exposure|Exposure_agent:Aflatoxin|Multiply|0.95-1|0-5% reduction of exposure based on 10 % more testing
| |
| Scenario|Action|exposure|Exposure_agent:Lead|Multiply|0.963-0.975|2.5-3.7% reduction of exposure based on more testing
| |
| Threshold|Yes|ERF|Exposure_agent:Lead;Observation:Threshold|Identity||Lanphear 2005 had the lowest exposure level at 24 ug/l
| |
| Threshold|No|ERF|Exposure_agent:Lead;Observation:Threshold|Replace|0-24|Sensitivity analysis looks whether reducing or removing threshold is important
| |
| Scenario|BAU|exposure||Identity||For completion
| |
| Scenario|Action|PAF|Exposure_agent:Noro virus|Multiply|0.65 : 1 : 1|35 % are from food industry so that is the max reduction from testing
| |
| Scenario|Action|PAF|Exposure_agent:Vegetables|Multiply|0.85 - 1|Based on RUORI modelling (see code)
| |
| Scenario|Action|PAF|Exposure_agent:Fruits|Multiply|0.82 - 1|Based on RUORI modelling (see code)
| |
| Adjust|BAU|exposure||Add|0.01|Roughly 98 % of all exposures for Exposure_agents is > 0.005. This prevents NaN in log scaling.
| |
| Scenario|Action|exposure|Exposure: To child; Age: Age 1|Replace|0.98 (0.17 - 2.9)|Based on RUORI modelling (see code)
| |
| Scenario|Action|exposure|Exposure: To eater; Age: Female 18-45|Replace|2.79 (0.56 - 7.08)|Based on RUORI modelling (see code)
| |
| Scenario|Action|exposure|Exposure: To eater; Age: Non female 18-45|Replace|11.38 (3.44 - 46.22)|Based on RUORI modelling (see code)
| |
| </t2b>
| |
| | |
| <t2b name="CollapseMarginals" index="Variable,Index,Probs,Function" obs="Dummy" desc="Description" unit="-">
| |
| BoD|incidenceSource,disabilityweightSource,populationSource,BoDSource||sum|1|Remove redundant
| |
| PAF|Unit, Exposure, Scaling,Exposcen, ER_function, ERFchoiceSource, exposureSource, bgexposureSource, BWSource, doseSource, thresholdSource, ERFSource, RRSource, frexposedSource, incidenceSource, InpPAFSource||sum|1|Remove redundant
| |
| case_burden|case_burdenSource||sum|1|Fill missing Ages
| |
| BoDattr|PAFSource, Hepatitis, Adjust||sum|1|Remove redundant
| |
| amount|assumpSource, oftenSource, muchSource, oftensideSource, muchsideSource, amountRawSource, effinfoSource, effrecommSource, amountSource||sum|1|Remove redundant
| |
| expo_indir|f_ingSource, t0.5Source,f_mtocSource, BFSource||sum|1|Remove redundant
| |
| dose|Source, concSource, expo_dirSrouce, exposureSource, BWSource, Source.1||sum|1|Remove redundant
| |
| conc|Fish|0, 1, 0, 0, 0, 0, 0.19, 0.19, 0.19, 0, 0, 0, 0, 0, 0, 0, 0.29, 0.14|sample|1|Probs relative to consumption; Baltic herring has equal weight with others combined because it is another scenario. Arctic char, Baltic herring, Bream, Burbot, Cod, Flounder, Perch, Pike, Pike-perch, Rainbow trout, River lamprey, Roach, Salmon, Sea trout, Sprat, Trout, Vendace, Whitefish
| |
| ERF|Exposure, Age||sum|1|Remove redundant
| |
| threshold|Exposure, Age||sum|1|Remove redundant
| |
| </t2b>
| |
| }}
| |
| | |
| === Laskenta ===
| |
| | |
| [[image:Terveysvaikutusten arviointimalli.svg|thumb|400px|Yleiskuva terveysvaikutusten laskentamallista.]]
| |
| | |
| ==== Keskeiset tulosteet ====
| |
| | |
| * Malliajo 5000 iteraatiota 11.8.2019 [http://fi.opasnet.org/fi-opwiki/index.php?title=Toiminnot:RTools&id=IMy5dmICIwDKkesL]
| |
| | |
| <rcode label="Laske uudestaan mallin perustulokset" graphics=1>
| |
| # This is code Op_fi5889/ on page [[Ruori]]
| |
| library(OpasnetUtils)
| |
| library(ggplot2)
| |
| library(thlGraphs)
| |
| library(plotly)
| |
| | |
| openv.setN(10)
| |
| | |
| objects.latest("Op_fi5889", code_name="model")
| |
| | |
| # First empty all objects for a fresh start. Otherwise may be problems with CheckDecisions.
| |
| oempty(all=TRUE)
| |
| | |
| InpBoD <- EvalOutput(InpBoD)
| |
| InpPAF <- EvalOutput(InpPAF)
| |
| utility <- EvalOutput(utility, verbose=TRUE)
| |
| | |
| # Sample from default and sensitivity scenario about lead threshold.
| |
| | |
| cat("Elintarvikeperäisen lyijyn vaikutukset herkkyystarkastelussa.\n")
| |
| oprint(summary(BoDattr[BoDattr$Exposure_agent=="Lyijy",]))
| |
| | |
| BoDattr <- CollapseMarginal(BoDattr,"Threshold","sample")
| |
| utility <- CollapseMarginal(utility,"Threshold","sample")
| |
| | |
| levels(BoDattr$Exposure_agent)[levels(BoDattr$Exposure_agent)=="Vihannesvaje"] <- "Kasvisvaje"
| |
| | |
| #levels(BoDattr$Response)
| |
| #[1] "Cancer morbidity" "IQ loss" "Listeriosis" "Liver cancer"
| |
| #[5] "Noro infection" "Sperm concentration" "Toxoplasmosis" "Yes or no dental defect"
| |
| #[9] "CHD death" "Diet high in sodium" "Diet low in fruits" "Diet low in vegetables"
| |
| | |
| levels(BoDattr$Response) <- c(
| |
| "Syöpä",
| |
| "Älykkyysosamäärän lasku",
| |
| "Listerioosi",
| |
| "Maksasyöpä",
| |
| "Noroinfektio",
| |
| "Miehen hedelmättömyys",
| |
| "Toksoplasmoosi",
| |
| "Hammasvaurio",
| |
| "Sydäntauti",
| |
| "Liika suola",
| |
| "Hedelmävaje",
| |
| "Kasvisvaje"
| |
| )
| |
| | |
| cat("Elintarvikeperäisiä tautitaakkoja Suomessa arpoen lyijylle oletetun tai matalamman kynnysarvon.\n")
| |
| tmp <- summary(oapply(BoDattr[BoDattr$Scenario=="BAU",],NULL,sum,c("Age","Response")))
| |
| oprint(data.frame(
| |
| Altiste = tmp$Exposure_agent,
| |
| Keskiarvo = signif(tmp$mean,2),
| |
| "95 luottamusväli" = paste0(signif(tmp$Q0.025,2)," - ", signif(tmp$Q0.975,2)),
| |
| Keskihajonta = signif(tmp$sd,2)
| |
| )[rev(match(lev, tmp$Exposure_agent)),])
| |
| | |
| cat("Elintarvikeperäisiä tautitaakkoja Suomessa ikä- ja tautiryhmittäin arpoen lyijylle oletetun tai matalamman kynnysarvon.\n")
| |
| tmp <- summary(BoDattr[BoDattr$Scenario=="BAU",])
| |
| oprint(data.frame(
| |
| Altiste = tmp$Exposure_agent,
| |
| Ikä = tmp$Age,
| |
| Vaste = tmp$Response,
| |
| Keskiarvo = signif(tmp$mean,2),
| |
| Mediaani = signif(tmp$median,2),
| |
| "95 luottamusväli" = paste0(signif(tmp$Q0.025,2)," - ", signif(tmp$Q0.975,2)),
| |
| Keskihajonta = signif(tmp$sd,2)
| |
| ))
| |
| | |
| cat("Ruori-skenaarioiden vaikutus tautitaakkaan\n")
| |
| tmp <- summary(utility)
| |
| oprint(data.frame(
| |
| Altiste = tmp$Exposure_agent,
| |
| Keskiarvo = signif(tmp$mean,2),
| |
| "95 luottamusväli" = paste0(signif(tmp$Q0.025,2)," - ", signif(tmp$Q0.975,2)),
| |
| Keskihajonta = signif(tmp$sd,2)
| |
| )[rev(match(lev, tmp$Exposure_agent)),])
| |
| | |
| dodge <- position_dodge(width=0.7)
| |
| | |
| if(FALSE) {
| |
|
| |
| gg <- ggplot(summary(oapply(BoDattr[BoDattr$Scenario=="BAU",],NULL,sum,"Age")),
| |
| aes(x=Exposure_agent, weight=unlist(mean), fill=Response))+geom_bar()+
| |
| theme(legend.position = "bottom")+
| |
| labs(
| |
| title="Elintarvikkeiden tautitaakkoja Suomessa",
| |
| subtitle="Haittapainotettua elinvuotta vuodessa (DALY/a)")+
| |
| coord_flip()
| |
|
| |
| print(gg)
| |
|
| |
| gg <- ggplot(summary(oapply(BoDattr, NULL, sum,c("Age","Response"))),
| |
| aes(x=Exposure_agent, weight=unlist(mean), fill=Scenario))+geom_bar(position="dodge")+
| |
| coord_flip(ylim=c(0,70000))+
| |
| labs(
| |
| title="Elintarvikeperäisiä tautitaakkoja Suomessa",
| |
| subtitle="Haittapainotettua elinvuotta vuodessa (DALY/a)")+
| |
| geom_errorbar(aes(ymin=unlist(Q0.025),ymax=unlist(Q0.975),group=Scenario),position=dodge, width=0.3)+
| |
| geom_text(aes(label=signif(unlist(mean),2), y=unlist(Q0.975)+5000, group=Scenario), position=dodge)
| |
|
| |
| print(gg)
| |
|
| |
| # Utility of actions
| |
|
| |
| gg <- ggplot(summary(utility),aes(x=Exposure_agent, weight=unlist(mean)))+geom_bar(fill="lightblue")+
| |
| coord_flip(ylim=c(-9000,0))+
| |
| labs(
| |
| title="Ruori-skenaarioiden vaikutus tautitaakkaan",
| |
| subtitle="Haittapainotettua elinvuotta vuodessa (DALY/a)")+
| |
| geom_errorbar(aes(ymin=unlist(Q0.025),ymax=unlist(Q0.975)), width=0.3)+
| |
| geom_text(aes(label=signif(unlist(mean),2), y=unlist(Q0.025)-600))
| |
|
| |
| print(gg)
| |
|
| |
| } else {
| |
|
| |
| ###### RUN THESE ON OWN COMPUTER WITH thlGraphs PACKAGE
| |
|
| |
| # levels(BoDattr$Exposure_agent)
| |
| # [1] "Aflatoksiini" "Dioksiini" "Norovirus" "Toksoplasma"
| |
| # [5] "Lyijy" "Listeria" "Tyydyttynyt rasva" "Vihannesvaje"
| |
| # [9] "Suola" "Hedelmävaje"
| |
|
| |
| levels(BoDattr$Exposure_agent) <- c(
| |
| "Aflatoxin","Dioxin","Noro virus","Toxoplasma", "Lead","Listeria",
| |
| "Saturated fat","Lack of vegetables","Sodium","Lack of fruits")
| |
|
| |
| thlBarPlot(summary(oapply(BoDattr[BoDattr$Scenario=="BAU",],NULL,sum,"Age")),
| |
| xvar=Exposure_agent, yvar=unlist(mean), groupvar=Response, legend.position="bottom",
| |
| colors=thlColors(n=12),
| |
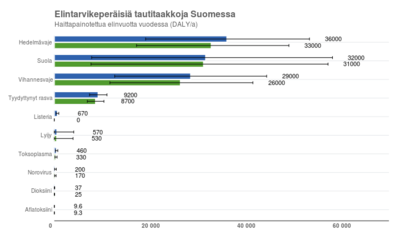
| title="Elintarvikkeiden tautitaakkoja Suomessa", subtitle="Haittapainotettua elinvuotta vuodessa (DALY/a)")+coord_flip()
| |
|
| |
| thlBarPlot(summary(oapply(BoDattr, NULL, sum,c("Age","Response"))),xvar=Exposure_agent, yvar=unlist(mean),
| |
| groupvar=Scenario,stacked=FALSE, ylimits=c(0,70000),
| |
| # title="Elintarvikeperäisiä tautitaakkoja Suomessa",
| |
| # subtitle="Haittapainotettua elinvuotta vuodessa (DALY/a)")+
| |
| title="Burden of disease of selected food-mediated risk factors in Finland",
| |
| subtitle="Disability-adjusted life years per year (DALY/a)")+
| |
| coord_flip()+
| |
| geom_errorbar(aes(ymin=unlist(Q0.025),ymax=unlist(Q0.975),group=Scenario),position=dodge, width=0.3)+
| |
| geom_text(aes(label=signif(unlist(mean),2), y=unlist(Q0.975)+5000, group=Scenario), position=dodge)
| |
|
| |
| ggsave("Ruori Burden of disease.png",width=10,height=6)
| |
| # ggsave("Ruori-tautitaakka.png",width=10,height=6)
| |
|
| |
|
| |
| # Error bars are not used in this plotly because it is unclear what it means in a stacked bar.
| |
| p <-plot_ly(
| |
| summary(oapply(BoDattr[BoDattr$Scenario=="BAU",], NULL, sum,c("Age"))),
| |
| y=~Exposure_agent,
| |
| x=~signif(mean,3),
| |
| text=~Response,
| |
| name=~Response,
| |
| type="bar",
| |
| orientation = "h"
| |
| ) %>%
| |
| layout(
| |
| barmode="stack",
| |
| title="Elintarvikeperäisiä tautitaakkoja Suomessa",
| |
| xaxis=list(title="Haittapainotettua elinvuotta vuodessa (DALY/a)"),
| |
| yaxis=list(title="")
| |
| )
| |
|
| |
| # pushIndicatorGraph(p, 117)
| |
| # ggsave("Ruori-tautitaakat.png",width=10/1.2,height=6/1.2)
| |
|
| |
| # Utility of actions
| |
|
| |
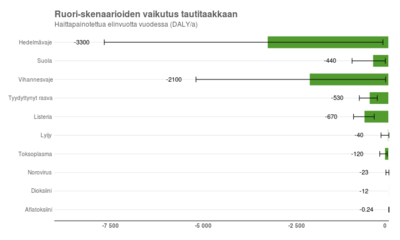
| thlBarPlot(summary(utility),xvar=Exposure_agent, yvar=unlist(mean), ylimits=c(-9000,0),
| |
| title="Ruori-skenaarioiden vaikutus tautitaakkaan",
| |
| subtitle="Haittapainotettua elinvuotta vuodessa (DALY/a)")+coord_flip()+
| |
| geom_errorbar(aes(ymin=unlist(Q0.025),ymax=unlist(Q0.975)), width=0.3)+
| |
| geom_text(aes(label=signif(unlist(mean),2), y=unlist(Q0.025)-600))
| |
|
| |
|
| |
| # ggsave("Ruori-toimenpideiden vaikutus.png",width=10/1.2,height=6/1.2)
| |
| }
| |
| </rcode>
| |
| | |
| ==== Arviointimallin alustus ====
| |
| | |
| * Linkit vanhempiin ajoihin [http://fi.opasnet.org/fi-opwiki/index.php?title=Ruori&oldid=35883#Arviointimallin_alustus arkistosta].
| |
| * Malliajo 10.8.2019 toksoplasma korjattu. Stored-versio [http://fi.opasnet.org/fi-opwiki/index.php?title=Toiminnot:RTools&id=VXNKkIPPZVSDhjUm]
| |
| | |
| <rcode name="model" label="Alusta koko Ruori-malli" graphics=1>
| |
| # This is code Op_fi5889/model on page [[Ruori]]
| |
| library(OpasnetUtils)
| |
| library(ggplot2)
| |
| | |
| # First empty all objects for a fresh start. Otherwise may be problems with CheckDecisions.
| |
| oempty(all=TRUE)
| |
| | |
| openv.setN(1000)
| |
| | |
| dat <- opbase.data("Op_fi5889", subset="Malliparametrit")[-1]
| |
| | |
| dec <- opbase.data("Op_fi5889", subset="Decisions")[-1]
| |
| DecisionTableParser(dec)
| |
| | |
| CTable <- opbase.data("Op_fi5889",subset="CollapseMarginals")
| |
| for(i in 1:ncol(CTable)) {CTable[[i]] <- as.character(CTable[[i]])}
| |
| CollapseTableParser(CTable)
| |
| | |
| cat("Laskennassa käytetty data.\n")
| |
| oprint(dat)
| |
| cat("Tarkastellut päätökset.\n")
| |
| oprint(dec)
| |
| cat("Aggregoidut marginaalit.\n")
| |
| oprint(CTable)
| |
| | |
| #' prepare adjusts the data table for ovariables. Requires function subgrouping from code Op_en2031/initiate on page [[Exposure-response function]]
| |
| #' @param dat data.frame
| |
| #' @param type type of data that is used. Must match content in column Type
| |
| #' @param drop columns to remove
| |
| #' @return data.frame
| |
| | |
| prepare <- function(dat, type=NULL, drop=NULL) {
| |
| out <- dat
| |
| if(!is.null(type)) out <- out[out$Type %in% type , ]
| |
| if(!is.null(drop)) out <- out[!colnames(out) %in% drop]
| |
| return(subgrouping(out))
| |
| }
| |
| | |
| objects.latest("Op_en2031", code_name="subgrouping") # [[Exposure-response function]] subgrouping
| |
| | |
| population <- Ovariable("population", data = prepare(dat,"population",c("Type","Exposure_agent","Response","Unit")))
| |
| | |
| exposure <- Ovariable("exposure", data = prepare(dat, "exposure", c("Type","Response")))
| |
| | |
| frexposed <- Ovariable("frexposed", data=prepare(dat, "frexposed", c("Type","Response","Unit")))
| |
| | |
| incidence <- Ovariable("incidence", data = prepare(dat,"incidence",c("Type","Exposure_agent","Unit")))
| |
| | |
| ERFChoice <- Ovariable(
| |
| "ERFchoice",
| |
| data=data.frame(
| |
| Response=c("IQ loss","Liver cancer","Yes or no dental defect","Cancer morbidity","Sperm concenctration"),
| |
| Exposure_agent=c("Lead","Aflatoxin","TEQ","TEQ","TEQ"),
| |
| Result=1)
| |
| )
| |
| | |
| case_burden <- Ovariable("case_burden", data= prepare(dat,"case burden",c("Type","Exposure_agent","Unit")))
| |
| | |
| InpPAF <- EvalOutput(Ovariable("InpPAF", data = prepare(dat,"PAF","Type")))
| |
| | |
| InpBoD <- EvalOutput(Ovariable("InpBoD", data = prepare(dat, "BoD", c("Type","Exposure_agent"))))
| |
| | |
| objects.latest("Op_en2261",code_name="BoDattr2") # [[Health impact assessment]]
| |
| | |
| #levels(BoDattr$Exposure_agent)
| |
| #[1] "Aflatoxin" "Lead" "TEQ" "Fruits" "Listeria" "Noro virus" "Saturated fat" "Sodium"
| |
| #[8] "Toxoplasma gondii" "Vegetables"
| |
| | |
| BoDattrOrigFormula <- BoDattr@formula
| |
| | |
| BoDattr@formula <- function(...) {
| |
| BoDattr <- BoDattrOrigFormula()
| |
|
| |
| levels(BoDattr$Exposure_agent) <- c("Aflatoksiini","Lyijy", "Dioksiini", "Hedelmävaje","Listeria","Norovirus",
| |
| "Tyydyttynyt rasva","Suola", "Toksoplasma","Vihannesvaje")
| |
|
| |
| lev <- oapply(BoDattr[BoDattr$Scenario=="BAU",],"Exposure_agent",sum)
| |
| lev <- lev$Exposure_agent[order(result(lev))]
| |
| BoDattr$Exposure_agent <- factor(BoDattr$Exposure_agent, levels=lev)
| |
| return(BoDattr)
| |
| }
| |
| | |
| utility <- Ovariable(
| |
| "utility",
| |
| dependencies = data.frame(Name="BoDattr"),
| |
| formula = function(...) {
| |
| out <- BoDattr * Ovariable(data=data.frame(Scenario=c("Action","BAU"),Result=c(1,-1)))
| |
| out <- oapply(out, cols=c("Scenario","Response","Age"),FUN=sum)
| |
| return(out)
| |
| }
| |
| )
| |
| | |
| utility <- EvalOutput(utility)
| |
| | |
| lev <- levels(BoDattr$Exposure_agent)
| |
| | |
| cat("exposure\n")
| |
| oprint(summary(exposure,marginals=c("Exposure_agent","Age","Scenario")),digits=7)
| |
| oprint(exposure@output[exposure$Iter==1,])
| |
| cat("dose\n")
| |
| oprint(summary(dose),digits=7)
| |
| cat("ERF\n")
| |
| oprint(ERF@output[ERF$Iter==1,],digits=7)
| |
| cat("incidence\n")
| |
| oprint(summary(incidence),digits=7)
| |
| cat("frexposed\n")
| |
| oprint(summary(frexposed),digits=7)
| |
| cat("PAF\n")
| |
| oprint(summary(PAF),digits=7)
| |
| cat("BoD\n")
| |
| oprint(summary(BoD),digits=7)
| |
| cat("BoDattr\n")
| |
| oprint(summary(BoDattr,marginals=c("Response","Exposure_agent","Scenario","Age")),digits=7)
| |
| cat("Vähennyspotentiaali\n")
| |
| tmp <- summary(utility, marginals=c("Exposure_agent"))
| |
| oprint(tmp[order(unlist(tmp$mean)),])
| |
| | |
| objects.store(list=setdiff(ls(), "wiki_username"))
| |
| cat("Objects", setdiff(ls(), "wiki_username"), "stored.\n")
| |
| | |
| ###################
| |
| # Disease burden
| |
| | |
| # Sample from default and sensitivity scenario about lead threshold.
| |
| | |
| cat("Elintarvikeperäisen lyijyn vaikutukset herkkyystarkastelussa.\n")
| |
| oprint(summary(BoDattr[BoDattr$Exposure_agent=="Lyijy",]))
| |
| | |
| BoDattr <- CollapseMarginal(BoDattr,"Threshold","sample")
| |
| utility <- CollapseMarginal(utility,"Threshold","sample")
| |
| | |
| cat("Elintarvikeperäisiä tautitaakkoja Suomessa\n")
| |
| tmp <- summary(oapply(BoDattr[BoDattr$Scenario=="BAU",],NULL,sum,c("Age","Response")))
| |
| oprint(data.frame(
| |
| Altiste = tmp$Exposure_agent,
| |
| Keskiarvo = signif(tmp$mean,2),
| |
| "95 luottamusväli" = paste0(signif(tmp$Q0.025,2)," - ", signif(tmp$Q0.975,2)),
| |
| Keskihajonta = signif(tmp$sd,2)
| |
| )[rev(match(lev, tmp$Exposure_agent)),])
| |
| | |
| cat("Ruori-skenaarioiden vaikutus tautitaakkaan\n")
| |
| tmp <- summary(utility)
| |
| oprint(data.frame(
| |
| Altiste = tmp$Exposure_agent,
| |
| Keskiarvo = signif(tmp$mean,2),
| |
| "95 luottamusväli" = paste0(signif(tmp$Q0.025,2)," - ", signif(tmp$Q0.975,2)),
| |
| Keskihajonta = signif(tmp$sd,2)
| |
| )[rev(match(lev, tmp$Exposure_agent)),])
| |
| | |
| dodge <- position_dodge(width=0.7)
| |
| | |
| ggplot(summary(oapply(BoDattr[BoDattr$Scenario=="BAU",],NULL,sum,"Age")),
| |
| aes(x=Exposure_agent, weight=unlist(mean), fill=Response))+geom_bar()+
| |
| theme(legend.position = "bottom")+
| |
| labs(
| |
| title="Elintarvikkeiden tautitaakkoja Suomessa",
| |
| subtitle="Haittapainotettua elinvuotta vuodessa (DALY/a)")+
| |
| coord_flip()
| |
| | |
| ggplot(summary(oapply(BoDattr, NULL, sum,c("Age","Response"))),
| |
| aes(x=Exposure_agent, weight=unlist(mean), fill=Scenario))+geom_bar(position="dodge")+
| |
| coord_flip(ylim=c(0,70000))+
| |
| labs(
| |
| title="Elintarvikeperäisiä tautitaakkoja Suomessa",
| |
| subtitle="Haittapainotettua elinvuotta vuodessa (DALY/a)")+
| |
| geom_errorbar(aes(ymin=unlist(Q0.025),ymax=unlist(Q0.975),group=Scenario),position=dodge, width=0.3)+
| |
| geom_text(aes(label=signif(unlist(mean),2), y=unlist(Q0.975)+5000, group=Scenario), position=dodge)
| |
| | |
| # Utility of actions
| |
| | |
| ggplot(summary(utility),aes(x=Exposure_agent, weight=unlist(mean)))+geom_bar(fill="lightblue")+
| |
| coord_flip(ylim=c(-9000,0))+
| |
| labs(
| |
| title="Ruori-skenaarioiden vaikutus tautitaakkaan",
| |
| subtitle="Haittapainotettua elinvuotta vuodessa (DALY/a)")+
| |
| geom_errorbar(aes(ymin=unlist(Q0.025),ymax=unlist(Q0.975)), width=0.3)+
| |
| geom_text(aes(label=signif(unlist(mean),2), y=unlist(Q0.025)-600))
| |
| </rcode>
| |
| | |
| ==== Dioksiini- ja muu valmistelu ====
| |
| | |
| * Linkit vanhempiin ajoihin löytyvät [http://fi.opasnet.org/fi-opwiki/index.php?title=Ruori&oldid=35883#Dioksiiniskenaario arkistosta].
| |
| * Malliajo 18.6.2019 toimii omalla koneella [http://fi.opasnet.org/fi-opwiki/index.php?title=Toiminnot:RTools&id=PRqtiUhfxvL0V8hK]
| |
| | |
| <rcode name="prepare" graphics=1>
| |
| # This is code Op_fi5889/prepare on page [[Ruori]]
| |
| | |
| library(OpasnetUtils)
| |
| library(ggplot2)
| |
| | |
| rm(list=ls())
| |
| rm(list=ls(envir = openv),envir=openv)
| |
| | |
| openv.setN(10000)
| |
| | |
| ##############################
| |
| | |
| # Pb exposure in children
| |
| # Data feched from \\helfs01.thl.fi/documents/YMAL/Projects/TUORI/tautitaakka/lyijy/Lyijy_tautitaakkadata.xlsx
| |
| | |
| # Pb <- re#ad.csv("clipboard",sep="\t",dec=",")
| |
| # ggplot(Pb, aes(x=Pb, fill=as.character(Age)))+geom_density(alpha=0.5)
| |
| | |
| | |
| # Population data
| |
| | |
| if(FALSE) {
| |
| # Read population data 2018 from Statistics Finland
| |
| #vae <- re#ad.csv("https://pxnet2.stat.fi:443/PXWeb/sq/ac3373d0-e303-4c67-b32a-73c6d26df809", skip=2)
| |
| #vae$Ikä <- as.numeric(gsub(" -","",as.character(vae$Ikä)))
| |
|
| |
| cat("Ages 1, 25-64, 65-74, 25-29, 70+, Total population, 0, 1+, Female 18-45, Non female 18-45\n")
| |
| c(
| |
| sum(vae$X2018[vae$Ikä==1]), # Age 1
| |
| sum(vae$X2018[vae$Ikä>=25 & vae$Ikä <65]), # Age 25-64
| |
| sum(vae$X2018[vae$Ikä>=65 & vae$Ikä <75]), # Age 65-74
| |
| sum(vae$X2018[vae$Ikä>=25 & vae$Ikä <70]), # Age 25-69
| |
| sum(vae$X2018[vae$Ikä>=70 & vae$Ikä <101]), # Age 70+
| |
| sum(vae$X2018), # Total population
| |
| sum(vae$X2018[vae$Ikä==0]), # Age 0
| |
| sum(vae$X2018[vae$Ikä>=1]), # Age 1+
| |
| sum(vae$X2018[vae$Ikä>=18 & vae$Ikä <46 & vae$Sukupuoli=="Naiset"]), #Female 18-45
| |
| sum(vae$X2018[!(vae$Ikä>=18 & vae$Ikä <46 & vae$Sukupuoli=="Naiset")]) #Non female 18-45
| |
| )
| |
| }
| |
| | |
| ##########################3
| |
| | |
| dat <- opbase.data("Op_fi5889", subset="Malliparametrit")[-1]
| |
| | |
| dec <- opbase.data("Op_fi5889", subset="Decisions")[-1]
| |
| DecisionTableParser(dec)
| |
| | |
| CTable <- opbase.data("Op_fi5889",subset="CollapseMarginals")
| |
| for(i in 1:ncol(CTable)) {CTable[[i]] <- as.character(CTable[[i]])}
| |
| CollapseTableParser(CTable)
| |
| | |
| cat("Laskennassa käytetty data.\n")
| |
| oprint(dat)
| |
| cat("Tarkastellut päätökset.\n")
| |
| oprint(dec)
| |
| cat("Aggregoidut marginaalit.\n")
| |
| oprint(CTable)
| |
| | |
| #' prepare adjusts the data table for ovariables. Requires function subgrouping from code Op_en2031/initiate on page [[Exposure-response function]]
| |
| #' @param dat data.frame
| |
| #' @param type type of data that is used. Must match content in column Type
| |
| #' @param drop columns to remove
| |
| #' @return data.frame
| |
| | |
| prepare <- function(dat, type=NULL, drop=NULL) {
| |
| out <- dat
| |
| if(!is.null(type)) out <- out[out$Type %in% type , ]
| |
| if(!is.null(drop)) out <- out[!colnames(out) %in% drop]
| |
| return(subgrouping(out))
| |
| }
| |
| | |
| objects.latest("Op_en2031", code_name="initiate") # [[Exposure-response function]] subgrouping
| |
| | |
| ###### Concentration data
| |
| | |
| objects.latest("Op_en3104", code_name="preprocess") # [[EU-kalat]]") eu2
| |
| | |
| # Get a lognormal concentration distribution for each fish species using same sd and individual means
| |
| | |
| conc <- Ovariable(
| |
| "conc",
| |
| dependencies=data.frame(Name=c("eu2")),
| |
| formula = function(...) {
| |
| out <- (oapply(eu2[eu2$Compound %in% c("PCDDF", "PCB") , ], NULL, sum, "Compound"))
| |
| out <- oapply(log(out), c("Fish","eu2Source"), mean)
| |
| result(out) <- paste(result(out), oapply(out, c("eu2Source"), sd)$eu2Result, sep="+-")
| |
| out <- out@output[colnames(out@output)!="eu2Source"]
| |
| colnames(out)[colnames(out)=="eu2Result"] <- "Result"
| |
| out$Exposure_agent <- "TEQ"
| |
| out <- exp(EvalOutput(Ovariable("conc",data=out)))
| |
| out$Scenario <- ifelse(out$Fish=="Baltic herring", "BAU","Action")
| |
| return(out)
| |
| }
| |
| )
| |
| | |
| ######## Fish intake data
| |
| | |
| objects.latest("Op_en7749", code_name = "initiate") # [[Goherr: Fish consumption study]]
| |
| ## Variables assump, often, much, oftenside, muchside, amountRaw, effinfo, effrecomm, amount
| |
| | |
| effinfo <- 0 # We are not interested in changes in amount
| |
| effrecomm <- 0
| |
| | |
| amountOrigFormula <- amount@formula
| |
| amount@formula <- function(...) {
| |
| out <- amountOrigFormula(...)
| |
| out <- oapply(out[out$Fish=="Herring",], NULL, sum, "Fish")
| |
| out <- out * info
| |
| return(out)
| |
| }
| |
| | |
| # Stores non-marginal columns for further use.
| |
| info <- Ovariable(
| |
| "info",
| |
| dependencies = data.frame(Name = c("jsp")),
| |
| formula = function(...) {
| |
| out <- unique(jsp@output[c("Iter","Country","Gender","Ages","Row")])
| |
| out <- out[out$Country=="FI",]
| |
| out$Group <- paste(out$Gender, out$Ages)
| |
| out$Group <- ifelse(out$Group=="Female 18-45", out$Group, "Non female 18-45")
| |
| out$Result <- 1
| |
| return(out)
| |
| }
| |
| )
| |
| | |
| expo_bg <- 0
| |
| | |
| expo_dir <- Ovariable(
| |
| "expo_dir",
| |
| dependencies=data.frame(Name=c("amount","conc","expo_bg")),
| |
| formula = function(...) {
| |
| out <- amount * conc
| |
|
| |
| out <- Ovariable(data = data.frame(
| |
| Exposcen = c("BAU", "No exposure"),
| |
| Result = c(1, 0)
| |
| )) * out + expo_bg
| |
| out$Exposure <- as.factor(
| |
| ifelse(
| |
| out$Exposure_agent %in% c("DHA", "MeHg"),
| |
| "To child",
| |
| "To eater"
| |
| )
| |
| )
| |
| out@marginal[colnames(out@output)=="Exposcen"]<- TRUE
| |
| return(out)
| |
| }
| |
| )
| |
| | |
| ### mc2d makes a 2D Monte Carlo with assumed 50 individuals in data. Exposure is their average.
| |
| | |
| exposure <- Ovariable(
| |
| "exposure",
| |
| dependencies = data.frame(
| |
| Name = c(
| |
| "expo_dir", # direct exposure, i.e. the person eats or breaths the exposure agent themself
| |
| "expo_indir", # indirect exposure, i.e. the person (typically fetus or infant) is exposed via someone else (mother)
| |
| "mc2d" # 2D Monte Carlo function
| |
| ),
| |
| Ident = c(
| |
| NA,
| |
| "Op_en7797/expo_indir", # [[Infant's dioxin exposure]] # expo_indir
| |
| "Op_en7805/mc2d") # [[Two-dimensional Monte Carlo]]
| |
| ),
| |
| formula = function(...) {
| |
| out <- combine(expo_dir, expo_indir)
| |
| out <- unkeep(out, "Source.1", sources=TRUE)
| |
| out <- mc2d(out)
| |
| return(out)
| |
| }
| |
| )
| |
| exposure@meta <- c(
| |
| exposure@meta,
| |
| list(units = "To eater: pg /day; to child: pg /g fat")
| |
| )
| |
| | |
| mc2dparam<- list(
| |
| N2 = 1000, # Number of iterations in the new Iter
| |
| strength = 50, # Sample size to which the fun is to be applied. Resembles number of observations
| |
| run2d = TRUE, # Should the mc2d function be used or not?
| |
| info = info, # Ovariable that contains additional indices, e.g. newmarginals.
| |
| newmarginals = c("Group","Exposure"), # Names of columns that are non-marginals but should be sampled enough to become marginals
| |
| method = "bootstrap", # which method to use for 2D Monte Carlo? Currently bootsrap is the only option.
| |
| fun = mean # Function for aggregating the first Iter dimension.
| |
| )
| |
| | |
| exposure <- EvalOutput(exposure,verbose=TRUE)
| |
| | |
| oprint(summary(exposure[exposure$Exposcen=="BAU",], marginals=c("Exposure_agent","Scenario","Group","Exposure")))
| |
| oprint(summary(conc))
| |
| oprint(summary(amount*info,marginals="Group"))
| |
| | |
| ggplot(conc@output, aes(x=concResult, colour=Scenario))+stat_ecdf()+scale_x_log10()
| |
| | |
| ggplot((info*amount)@output, aes(x=amountResult+0.01, colour=Group))+stat_ecdf()+scale_x_log10()
| |
| | |
| ggplot(info*expo_indir@output, aes(x=expo_indirResult+0.01, colour=Group))+stat_ecdf()+scale_x_log10()+facet_grid(Group~Exposure)
| |
| | |
| ggplot(exposure@output[exposure$Exposcen=="BAU",], aes(x=exposureResult, colour=Scenario))+geom_density()+facet_grid(Group~Exposure)
| |
| | |
| tmp <- summary(exposure[exposure$Exposcen=="BAU",], marginals=c("Scenario","Group","Exposure"))
| |
| tmp[4:10] <- as.data.frame(lapply(tmp[4:10], function(x) round(x, 2)))
| |
| tmp$out <- paste0(tmp$mean, " (",tmp$Q0.025, " - ", tmp$Q0.975, ")")
| |
| oprint(tmp)
| |
| tmp$out
| |
| | |
| ##############################
| |
| | |
| # Domestic fish consumption. Used to give weights to fish species concentration data
| |
| | |
| tmp <- opbase.data("Op_en7749", subset="Fish consumption as food in Finland")
| |
| tmp <- tmp[tmp$Origin=="domestic fish" & !tmp$Species %in% c(
| |
| "Total", "Farmed rainbow trout","Baltic herring", "Other domestic fish") & tmp$Year==2017 ,
| |
| !colnames(tmp) %in% c("Obs","Origin", "Year")]
| |
| colnames(tmp)[colnames(tmp)=="Species"] <- "Fish"
| |
| levels(tmp$Fish)[
| |
| match(c("European whitefish", "Pike perch"), levels(tmp$Fish))] <- c("Whitefish","Pike-perch")
| |
| tmp$Result <- tmp$Result / sum(tmp$Result)
| |
| tmp <- merge(unique(eu2@output["Fish"]), tmp, all.x=TRUE)
| |
| tmp$Result[is.na(tmp$Result)] <- 0
| |
| tmp$Result[tmp$Fish=="Baltic herring"] <- 1 # Baltic herring is on BAU scenario and gets equal weight with others combined
| |
| | |
| cat("Copy these weights to CollapseMarginal table for conc Collapsing.\n")
| |
| round(tmp$Result,2)
| |
| | |
| ###### This is temporary code that is used to calculate the option "Action" of decision "Scenario".
| |
| | |
| population <- Ovariable("population", data = prepare(dat,"population",c("Type","Exposure_agent","Response","Unit")))
| |
| | |
| reduction <- Ovariable("reduction", data = prepare(dat,"reduction",c("Type","Response")))
| |
| | |
| intake <- Ovariable("intake", data = prepare(dat,"intake",c("Type","Response")))
| |
| | |
| recommendation <- Ovariable("recommendation", data = prepare(dat,"recommendation",c("Type","Response")))
| |
| | |
| eaters <- Ovariable("eaters", data = prepare(dat,"eaters",c("Type","Response","Unit")))
| |
| | |
| PAF_factor <- Ovariable(
| |
| "PAF_factor",
| |
| dependencies=data.frame(Name=c("reduction","intake","recommendation","eaters","population")),
| |
| formula = function(...) {
| |
| out <- 1 - reduction / (intake - recommendation) * eaters / population
| |
| return(out)
| |
| }
| |
| )
| |
| | |
| PAF_factor <- EvalOutput(PAF_factor)
| |
| | |
| ggplot(PAF_factor@output, aes(x=PAF_factorResult, fill=Exposure_agent))+geom_density()+facet_wrap(~Exposure_agent)
| |
| | |
| summary(PAF_factor)
| |
| | |
| # The PAF_factor distributions for saturated fat and sodium are NOT normally distributed. Instead, triangular
| |
| # distribution seems to be a reasonable fit with parameters:
| |
| # Sodium: triangular 0.975 : 0.990 : 0.993
| |
| # Saturated fat: triangular 0.911 : 0.944 : 0.971
| |
| | |
| ##### Q25 was used when 25 quantiles were estimated. Now we use rnorm estimate.
| |
| Q25 <- function(x) {
| |
| return(round(quantile(x, probs = seq(0.02, 0.98, 0.04)),1))
| |
| }
| |
| | |
| summary(exposure[exposure$Exposcen=="BAU",], marginals=c("Exposure","Group","Scenario"), "Q25")
| |
| | |
| ################ Insight network
| |
| | |
| gr <- scrape(type="assessment")
| |
| objects.latest("Op_en3861", "makeGraph") # [[Insight network]]
| |
| gr <- makeGraph(gr)
| |
| #export_graph(gr, "ruori.svg")
| |
| render_graph(gr)
| |
| </rcode>
| |
| | |
| ==== Maavertailu hygienian ja ravinnon tautitaakasta ====
| |
| | |
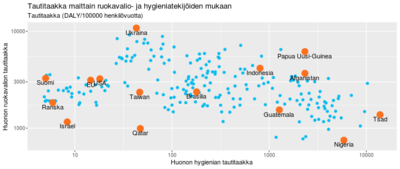
| Koodin ajamiseksi on ensin haettava [http://ghdx.healthdata.org/gbd-results-tool?params=gbd-api-2017-permalink data] IHME-instituutista. Koodi tuottaa kuvan {{#l:Tautitaakka maittain ruokavalio ja hygienia.svg}}.
| |
| | |
| <rcode graphics=1>
| |
| # This is code Op_fi5889& on page Ruori
| |
| | |
| library(OpasnetUtils)
| |
| library(plotly)
| |
| library(thlGraphs)
| |
| # permalink to IHME data: http://ghdx.healthdata.org/gbd-results-tool?params=gbd-api-2017-permalink/7c1842c34d51287572a49c78b74c4801
| |
| | |
| dat <- opasnet.csv(
| |
| "/d/d3/IHME-GBD_2017_diet_hygiene_by_country.zip", wiki="opasnet_en",
| |
| unzip="IHME-GBD_2017_DATA-c3ad9a2c-1.csv",
| |
| dec=".", sep=",", header=TRUE, quote="\""
| |
| )
| |
| | |
| tmp <- reshape(
| |
| dat[c("location_name","rei_name","val","upper","lower")],
| |
| v.names=c("val","upper","lower"),
| |
| timevar="rei_name",
| |
| idvar=c("location_name"),
| |
| direction="wide"
| |
| )
| |
| | |
| #colnames(tmp)
| |
| #[1] "location_name"
| |
| #[2] "val.Unsafe water, sanitation, and handwashing"
| |
| #[3] "upper.Unsafe water, sanitation, and handwashing"
| |
| #[4] "lower.Unsafe water, sanitation, and handwashing"
| |
| #[5] "val.Dietary risks"
| |
| #[6] "upper.Dietary risks"
| |
| #[7] "lower.Dietary risks"
| |
| | |
| colnames(tmp) <- c("Location","Hygiene","Hygupper","Hyglower","Diet","Dietupper","Dietlower")
| |
| | |
| eng <- c(
| |
| "Finland",
| |
| "Israel",
| |
| "France",
| |
| "Qatar",
| |
| "Taiwan",
| |
| "Indonesia",
| |
| "Guatemala",
| |
| "Papua New Guinea",
| |
| "European Union",
| |
| "United States",
| |
| "Ukraine",
| |
| "Afghanistan",
| |
| "Nigeria",
| |
| "Chad",
| |
| "Brazil"
| |
| )
| |
| fi <- c(
| |
| "Suomi",
| |
| "Israel",
| |
| "Ranska",
| |
| "Qatar",
| |
| "Taiwan",
| |
| "Indonesia",
| |
| "Guatemala",
| |
| "Papua Uusi-Guinea",
| |
| "EU",
| |
| "USA",
| |
| "Ukraina",
| |
| "Afganistan",
| |
| "Nigeria",
| |
| "Tsad",
| |
| "Brasilia"
| |
| )
| |
| | |
| tmp$Point <- tmp$Location %in% eng
| |
| tmp$Paikka <- fi[match(tmp$Location,eng)]
| |
| tmp$Diettext <- 0.9 * tmp$Diet
| |
| tmp$Diettext <- ifelse(tmp$Location=="United States", 1.25 * tmp$Diettext, tmp$Diettext)
| |
| | |
| p <- plot_ly() %>%
| |
| add_trace(
| |
| x=signif(tmp$Hygiene,3),
| |
| y=~signif(tmp$Diet,3),
| |
| text=tmp$Location,
| |
| name="all countries",
| |
| type="scatter",
| |
| mode="markers",
| |
| marker=list(color=rgb(10,187,239,maxColorValue = 255))
| |
| ) %>%
| |
| add_trace(
| |
| x=~signif(tmp$Hygiene[tmp$Point],3),
| |
| y=signif(tmp$Diet[tmp$Point],3),
| |
| text=tmp$Paikka[tmp$Point],
| |
| name="selected",
| |
| type="scatter",
| |
| mode="markers",
| |
| marker=list(size=10, color=rgb(251,112,29, maxColorValue = 255))
| |
| ) %>%
| |
| add_trace(
| |
| x=~signif(tmp$Hygiene[tmp$Point],3),
| |
| y=signif(0.9*tmp$Diet[tmp$Point],3),
| |
| text=tmp$Paikka[tmp$Point],
| |
| name="selected",
| |
| type="scatter",
| |
| mode="text",
| |
| hoverinfo="skip",
| |
| text=list(color=rgb(251,112,29, maxColorValue = 255))
| |
| ) %>%
| |
| layout(
| |
| xaxis = list(title="Huonon hygienian tautitaakka",type = "log"),
| |
| yaxis = list(title="Huonon ruokavalion tautitaakka",type = "log"),
| |
| title="Tautitaakka maittain ruokavalio- ja hygieniatekijöiden mukaan (DALY/100000 henkilövuotta)",
| |
| showlegend=FALSE
| |
| )
| |
| | |
| #pushIndicatorGraph(p, 191)
| |
| | |
| ggplot() +
| |
| geom_point(data=tmp, aes(x=Hygiene, y=Diet),color=rgb(10,187,239,maxColorValue = 255))+
| |
| geom_point(data=tmp[tmp$Point,], aes(x=Hygiene, y=Diet), color=rgb(251,112,29, maxColorValue = 255),size=4)+
| |
| geom_text(data=tmp[tmp$Point,], size=7, aes(x=Hygiene, y=Diettext, label=Paikka))+#Location))+
| |
| scale_x_log10() + scale_y_log10()+
| |
| thlTheme(x.axis.title = TRUE, base.size=32, y.axis.title.vertical = TRUE)+ # Assumes an adjustment available at github.com/jtuomist/thlGraph
| |
| labs(
| |
| title="Tautitaakka maittain ruokavalio- ja hygieniatekijöiden mukaan",
| |
| subtitle="Tautitaakka (DALY/100000 henkilövuotta)",
| |
| x="Huonon hygienian tautitaakka",
| |
| y="Huonon ruokavalion tautitaakka"
| |
| # title="Burden of disease of dietary and hygiene risk factors by country",
| |
| # subtitle="Burden of disease (DALY/100000 person-years",
| |
| # x="Burden of disease due to poor hygiene",
| |
| # y="Burden of disease due to dietary factors"
| |
| )
| |
| | |
| #ggsave(
| |
| # "Tautitaakka maittain ruokavalio ja hygienia.png",
| |
| ## "Burden of disease of diet and hygiene by country.png",
| |
| # width=9.7/0.6, height=4.1/0.6)
| |
| </rcode>
| |
| | |
| ==== Tautitaakkakuvia ====
| |
| | |
| Kuvat on tehty Ruori-seminaariin 25.4.2019. Uudemmat kuvat löytyvät tuloksista.
| |
| | |
| <rcode label="Ruori ja tautitaakka -esityksen kuvat (aja omalla koneella)">
| |
| # Tämä on koodi Op_fi5889 sivulla [[Ruori]]
| |
| | |
| library(OpasnetUtils)
| |
| library(tidyverse)
| |
| library(treemap)
| |
| library(plotly)
| |
| | |
| #objects.latest("Op_en6007", code_name = "hnh2035") # [[OpasnetUtils/Drafts]] pushIndicatorGraph
| |
| | |
| ##### Finnish translations
| |
| transl <- as_tibble(opbase.data("Op_fi3944", subset="Tautiluokittelu")) %>% # [[Tautitaakka Suomessa]]
| |
| mutate(Id=as.integer(as.character(Id)))
| |
| | |
| BS <- 24
| |
| | |
| palet <- c(
| |
| '#a6cee3','#1f78b4','#b2df8a','#33a02c','#fb9a99','#e31a1c','#fdbf6f',
| |
| '#ff7f00','#cab2d6','#6a3d9a','#ffff99','#b15928', # First 12 colours from Colorbrewer Paired.
| |
| 'cyan2','cyan4','plum1','plum4', 'darkslategray4','darkslategray1','firebrick3'
| |
| )
| |
| | |
| ###################### Graphs for the Tautitaakka auttaa hahmottamaan ... article
| |
| | |
| # DALYs by causes (risks not included)
| |
| dc <-
| |
| as_tibble(opasnet.csv("2/2d/IHME_Fin_Risks_by_Cause.zip", wiki="opasnet_en",
| |
| unzip="IHME-GBD_2017_DATA-8ce9adcf-1.csv",sep=",",dec=".",header=TRUE)) %>%
| |
| left_join(transl[transl$Type=="Cause",], by=c("cause_id"="Id")) %>%
| |
| mutate(cause_name = Result) %>% # transl combines some entries from cause_name to Name (in Finnish: Result)
| |
| # "Neglected tropical diseases and malaria",
| |
| # "HIV/AIDS and sexually transmitted infections",
| |
| # "Enteric infections" >> "Other infectious diseases"
| |
| filter(measure_name=="DALYs (Disability-Adjusted Life Years)" & metric_name=="Number" & year == 2017) %>%
| |
| group_by(cause_name) %>%
| |
| summarise(value=sum(val)) %>%
| |
| mutate(cause_name = factor(cause_name, levels=cause_name[order(value)])) %>%
| |
| mutate(valy = value/1000 + ifelse(value<250000, 20, -40)) # unit kDALY/a
| |
| | |
| plot_cause <- ggplot(dc2, aes(x=cause_name,weight=value/1000,fill=cause_name, label=round(value/1000)))+
| |
| geom_bar(position="stack")+geom_text(aes(y=valy))+coord_flip()+
| |
| guides(fill=FALSE)+
| |
| scale_fill_manual(values = rev(palet))+
| |
| labs(
| |
| title="Tautitaakka Suomessa syittäin 2017",
| |
| x="Tauti tai haitta",
| |
| y="Tautitaakka (tuhatta DALYa vuodessa)")
| |
| | |
| plot_cause
| |
| # ggsave("Tautitaakka Suomessa 2017.svg", width=8, height=5) # Png conversion: 300 pixels/inch, font size 4 times larger
| |
| | |
| ############## DALYs by causes and risks
| |
| | |
| dcr <- as_tibble(
| |
| opasnet.csv("2/2d/IHME_Fin_Risks_by_Cause.zip", wiki="opasnet_en",
| |
| unzip="IHME-GBD_2017_DATA-8c9ca17f-1.csv",sep=",",dec=".",header=TRUE)
| |
| ) %>%
| |
| left_join(transl[transl$Type=="Cause",], by=c("cause_id"="Id")) %>%
| |
| filter(measure_name=="DALYs (Disability-Adjusted Life Years)" & metric_name=="Number" & year == 2017) %>%
| |
| # mutate(cause_name = factor(Result, levels=dc2$cause_name)) %>% # transl combines some entries from cause_name to Name (in Finnish: Result)
| |
| left_join(transl[transl$Type=="Risk",], by=c("rei_id"="Id"))
| |
| | |
| dcr <- dcr %>%
| |
| mutate(
| |
| cause_name=factor(Result.x,levels=levels(dc$cause_name)),
| |
| rei_name=factor(Result.y, levels = aggregate(dcr["val"],dcr["Result.y"],sum) %>% transmute(Result.y[order(val)]) %>% pull())) %>%
| |
| group_by(rei_name, cause_name, Luokka.y) %>%
| |
| summarise(val = sum(val))
| |
| | |
| # mutate(rei_name = factor(rei_name, levels= dcr2$rei_name)) # transl combines some entries from cause_name to Name (in Finnish: Result)
| |
| | |
| # Strange discrepancy in two sources from IHME: especially in cardiovascular diseases.
| |
| # Reason not known, use dcr anyway; because errors not found.
| |
| # ihme <- read.csv("C:/Users/jtue/AppData/Local/Temp/download(1).csv")
| |
| # ihme <- ihme[ihme$Measure=="DALYs",]
| |
| # sum(ihme$Value)
| |
| # [1] 911630.9
| |
| # sum(dcr$val)
| |
| # [1] 1205234
| |
| # View(
| |
| # merge(
| |
| # aggregate(ihme["Value"], by=ihme["Cause.of.death.or.injury"], FUN=sum),
| |
| # aggregate(dcr["val"], by=dcr["cause_name"], FUN=sum),
| |
| # by.x=c("Cause.of.death.or.injury"), by.y=c("cause_name")
| |
| # )
| |
| # )
| |
| | |
| | |
| plot_risk <- ggplot(dcr, aes(x=rei_name, weight=val/1000, fill=cause_name))+geom_bar()+coord_flip()+
| |
| scale_fill_manual(values = rev(palet)[-8])+ # Skin diseases missing, so remove that colour
| |
| guides(fill = guide_legend(reverse=TRUE, title=NULL, keyheight=0.8))+
| |
| theme_gray(base_size=11)+ theme(legend.position=c(0.83,0.29)) +
| |
| labs(
| |
| title = "Tautitaakka Suomessa 2017 tunnettujen riskitekijöiden mukaan",
| |
| x="Riskitekijä", y="Tautitaakka (tuhatta DALYa vuodessa)")
| |
| | |
| plot_risk
| |
| # ggsave("Tautitaakka Suomessa 2017 riskitekijöittäin.svg", width=8, height=6) # Png conversion: 300 pixels/inch, font size 4 times larger
| |
| | |
| ruori <- data.frame(
| |
| Luokka.y = c(rep("Ympäristö",3),rep("Ravitsemus",4),rep("Ympäristö",3)),
| |
| rei_name = c(
| |
| "Aflatoksiini",
| |
| "Lyijy",
| |
| "Dioksiini",
| |
| "Tyydyttyneet rasvat",
| |
| "Suola",
| |
| "Hedelmien liian vähäinen saanti",
| |
| "Kasvisten liian vähäinen saanti",
| |
| "Norovirus",
| |
| "Listeria",
| |
| "Toksoplasma"
| |
| ),
| |
| val = c(
| |
| 8,
| |
| 6,
| |
| 22,
| |
| 8190,
| |
| 27062,
| |
| 35314,
| |
| 27778,
| |
| 1,
| |
| 660,
| |
| 432
| |
| ),
| |
| lower = c(
| |
| 6,
| |
| NA,
| |
| NA,
| |
| 5857,
| |
| 1330,
| |
| 20197,
| |
| 13740,
| |
| NA,
| |
| 330,
| |
| 270
| |
| ),
| |
| upper = c(
| |
| 13,
| |
| NA,
| |
| NA,
| |
| 11125,
| |
| 65542,
| |
| 53635,
| |
| 44716,
| |
| NA,
| |
| 880,
| |
| 702
| |
| )
| |
| )
| |
| | |
| dcr$rei_name <- as.character(dcr$rei_name)
| |
| | |
| # Subtract Ruori results from IHME background
| |
| dcr$val[dcr$rei_name=="Ravitsemusriskit" & dcr$cause_name=="Sydän- ja verisuonitaudit"] <-
| |
| dcr$val[dcr$rei_name=="Ravitsemusriskit" & dcr$cause_name=="Sydän- ja verisuonitaudit"] - sum(ruori$val[ruori$Luokka.y=="Ravitsemus"])
| |
| | |
| dcr$val[dcr$rei_name=="Muut ympäristöriskit" & dcr$cause_name=="Syöpä"] <-
| |
| dcr$val[dcr$rei_name=="Muut ympäristöriskit" & dcr$cause_name=="Syöpä"] - sum(ruori$val[ruori$Luokka.y=="Ympäristö"])
| |
| | |
| dcr <- rbind(dcr[colnames(dcr)!="cause_name"],ruori[!colnames(ruori) %in% c("upper","lower")])
| |
| dcr$rei_name[dcr$rei_name=="Ravitsemusriskit"] <- "Muut ravitsemusriskit"
| |
| | |
| pdf("Tautitaakka riskitekijöittäin.pdf", width=11.67, height=4.27)
| |
| treemap(dcr, index=c("Luokka.y","rei_name"),vSize="val",
| |
| title=paste0("Tunnettujen riskitekijöiden tautitaakka Suomessa 2017 (",
| |
| round(sum(dcr$val),-3)," DALY)"),
| |
| align.labels=list(c("center","top"),c("center","center")),
| |
| aspRatio = 2, border.col = "gray")
| |
| dev.off()
| |
| | |
| tmp <- ruori %>%
| |
| filter(rei_name %in% c("Aflatoksiini","Lyijy","Dioksiini","Norovirus","Listeria","Toksoplasma")) %>%
| |
| rename(Altiste=rei_name) %>%
| |
| mutate(Altiste = factor(Altiste, levels=ruori$rei_name[order(ruori$val)]))
| |
| | |
| thlBarPlot(tmp, xvar=Altiste, yvar=val)+
| |
| coord_flip()+geom_errorbar(data=tmp, aes(ymin=lower, ymax=upper), width=0.25, size=0.7)+
| |
| labs(
| |
| title="Ruorissa tutkittujen riskien tautitaakka",
| |
| subtitle = "Tautitaakka mikrobiologisille ja kemiallisille epäpuhtauksille (DALYa vuodessa)")
| |
| | |
| ggsave("Mikrobiologiset ja kemialliset riskit Ruorissa.svg", width=8, height=5)
| |
| | |
| tmp <- ruori %>%
| |
| filter(!rei_name %in% c("Aflatoksiini","Lyijy","Dioksiini","Norovirus","Listeria","Toksoplasma")) %>%
| |
| rename(Altiste=rei_name) %>%
| |
| mutate(Altiste = factor(Altiste, levels=ruori$rei_name[order(ruori$val)]))
| |
| | |
| thlBarPlot(tmp, xvar=Altiste, yvar=val)+
| |
| coord_flip()+geom_errorbar(data=tmp, aes(ymin=lower, ymax=upper), width=0.25, size=0.7)+
| |
| labs(
| |
| title="Ruorissa tutkittujen riskien tautitaakka",
| |
| subtitle = "Tautitaakka ravitsemusriskeille (DALYa vuodessa)")
| |
| | |
| ggsave("Ravitsemusriskit Ruorissa.svg", width=8, height=5)
| |
| | |
| ####################################
| |
| | |
| # Katso sivut [[:op_en:Goherr assessment]] ja [[:op_en:Health effects of Baltic herring and salmon: a benefit-risk assessment]]
| |
| objects.get('155401096341')
| |
| | |
| groups <- function(o) {
| |
| o$Group <- paste(o$Gender, o$Ages)
| |
| o$Group <- factor(o$Group, levels = c(
| |
| "Female 18-45",
| |
| "Male 18-45",
| |
| "Female >45",
| |
| "Male >45"
| |
| ))
| |
| return(o)
| |
| }
| |
| | |
| varit12 <- c(
| |
| '#2f62ad', # THL-värit, koko väripaletti. Nro 2
| |
| '#7cd0d8', # 4
| |
| '#571259', # 9
| |
| '#5faf2c', # 11
| |
| '#bf4073', # 14
| |
| '#3b007f', # 18
| |
| '#16994a', # 20
| |
| '#cccc72', # 22
| |
| '#0e1e47', # 3
| |
| '#25a5a2', # 5
| |
| '#cc7acc', # 7
| |
| '#244911' # 12
| |
| )
| |
| | |
| #### Burden of disease
| |
| | |
| # Figure 5. from Goherr assessment manuscript
| |
| | |
| if(FALSE) tmp <- groups(BoD * info) else tmp <- groups(BoDRaw * info)
| |
| | |
| tmp <- oapply(
| |
| tmp[!grepl("TWI", tmp$Resp) & tmp$Background=="Yes",],
| |
| INDEX=c("Resp","Group","Country","Cons.policy","Background"),
| |
| mean
| |
| )@output
| |
| | |
| # levels(tmp$Group)
| |
| # [1] "Female 18-45" "Male 18-45" "Female >45" "Male >45"
| |
| levels(tmp$Group) <- c("Naiset 18-45","Miehet 18-45","Naiset >45","Miehet >45")
| |
| | |
| #levels(tmp$Resp)
| |
| #[1] "Stroke" "Heart (CHD)" "Cancer" "Child's IQ" "Infertility"
| |
| #[6] "Tooth defect" "Dioxin TWI" "TWI 2018" "Vitamin D intake"
| |
| levels(tmp$Resp) <- c("Aivohalvaus","Sepelvaltimotauti","Syöpä","Lapsen ÄO","Hedelmättömyys",
| |
| "Hammasvaurio","","","D-vitamiini")
| |
| | |
| fig22 <- ggplot(tmp[tmp$Country=="FI" & tmp$Cons.policy=="BAU",],
| |
| aes(x=Group, weight=Result, fill=Resp))+
| |
| geom_bar()+facet_grid(Cons.policy ~ Country)+
| |
| theme_grey(base_size=BS-8)+
| |
| theme(
| |
| legend.position = "bottom"
| |
| # axis.text.x = element_text(angle = -90)
| |
| )+
| |
| labs(
| |
| title="Itämeren kalan aiheuttama tautitaakka Suomessa ryhmittäin",
| |
| y = "Tautitaakka (DALY/a)",
| |
| fill="",
| |
| x=""
| |
| )+
| |
| scale_fill_manual(values=varit12[-(7:8)])
| |
| | |
| fig22
| |
| ggsave("Itämeren kalan tautitaakka Suomessa.svg", width=8, height=5)
| |
| | |
| ######### Figure 6.
| |
| | |
| tmp <- groups(BoDRaw * info)
| |
| | |
| #> levels(tmp$Resp)
| |
| #[1] "Stroke" "Heart (CHD)" "Cancer" "Child's IQ" "Infertility"
| |
| #[6] "Tooth defect" "Dioxin TWI" "TWI 2018" "Vitamin D intake"
| |
| levels(tmp$Resp) <- c("Aivohalvaus","Sepelvaltimotauti","Syöpä","Lapsen ÄO","Hedelmättömyys",
| |
| "Hammasvaurio","TWI 2001","TWI 2018","D-vitamiini")
| |
| | |
| tmp@output <- rbind(
| |
| cbind(tmp@output,Focusgroup = "Kaikki"),
| |
| cbind(tmp@output[tmp$Group=="Female 18-45",],Focusgroup = "Nuoret naiset")
| |
| )
| |
| | |
| tmp <- oapply(
| |
| tmp[tmp$Cons.policy=="BAU" & tmp$Country=="FI",],
| |
| INDEX=c("Resp","Background","Focusgroup","Country"),
| |
| mean
| |
| )@output
| |
| | |
| tmp <- rbind(
| |
| cbind(
| |
| Objective="Kokonaisterveys, oletus",
| |
| tmp[tmp$Background=="Yes" & !grepl("TWI",tmp$Resp),]
| |
| ),
| |
| cbind(
| |
| Objective="Kokonaisterveys, taustatta",
| |
| tmp[tmp$Background=="No" & !grepl("TWI",tmp$Resp),]
| |
| ),
| |
| cbind(
| |
| Objective=tmp$Resp,
| |
| tmp
| |
| )[grepl("TWI",tmp$Resp),]
| |
| )
| |
| | |
| sums <- aggregate(tmp["Result"], by=tmp[c("Objective","Focusgroup","Country")],sum)
| |
| fig22b <- ggplot(tmp, aes(x=Objective, weight=Result))+
| |
| geom_bar(aes(fill=Resp))+facet_grid(Country ~ Focusgroup)+
| |
| theme_grey(base_size=BS-8)+
| |
| theme(
| |
| legend.position = "bottom",
| |
| axis.text.x = element_text(angle = -15, hjust=0.5)
| |
| )+
| |
| geom_text(data=sums, size=6, aes(label=round(Result,-2),y=pmax(200,Result+200)))+
| |
| labs(
| |
| title="Tautitaakka eri tavoitteiden näkökulmasta",
| |
| y = "Tautitaakka (DALY/a)",
| |
| fill="",
| |
| x=""
| |
| )+
| |
| scale_fill_manual(values=varit12)
| |
| fig22b
| |
| | |
| ggsave("Itämeren kalan tautitaakka eri näkökulmista.svg", width=10, height=7)
| |
| | |
| ############################ Health care costs
| |
| | |
| dat <- scrape.webtable(
| |
| "https://en.wikipedia.org/wiki/List_of_countries_by_total_health_expenditure_per_capita",
| |
| 6)
| |
| colnames(dat) <- c("Country","Y2000","Y2005","Y2010","Y2015")
| |
| dat$Y2015 <- as.numeric(gsub(",","",as.character(dat$Y2015)))
| |
| dat$Country <- as.character(dat$Country)
| |
| dat$Country[match(c(
| |
| "Iran (Islamic Republic of)",
| |
| "Lao People's Democratic Republic",
| |
| "United Republic of Tanzania",
| |
| "United States of America",
| |
| "Venezuela (Bolivarian Republic of)",
| |
| "Viet Nam"
| |
| ),dat$Country)] <- c("Iran","Laos","Tanzania","United States","Venezuela","Vietnam")
| |
| | |
| dat2 <- read.csv("C:/Users/jtue/AppData/Local/Temp/IHME-GBD_2017_DATA-96ba4b83-1.csv")
| |
| levels(dat2$location_name)[match(c(
| |
| "The Bahamas","The Gambia"),levels(dat2$location_name))] <- c("Bahamas","Gambia")
| |
| | |
| dat3 <- merge(dat,dat2, by.x="Country",by.y="location_name", all=TRUE)
| |
| dat3$label <- ifelse(dat3$Country %in% c("Finland","Sweden","United States","Poland"), dat3$Country, " ")
| |
| | |
| tmp <- dat3[dat3$metric_name=="Rate" & dat3$cause_name=="All causes",]
| |
| tmp$label[match(c("Finland","Sweden","United States","Poland"),tmp$label)] <- c("Suomi","Ruotsi", "USA","Puola")
| |
| | |
| ggplot(tmp, aes(x=Y2015,y=val/100000, label=label, colour=label))+
| |
| geom_point()+geom_text()+
| |
| labs(
| |
| title="Tautitaakka terveysmenojen funktiona maittain 2015",
| |
| y="Tautitaakka per henkilö (DALY)",
| |
| x="Terveydenhuoltomenot per henkilö (USD)"
| |
| )
| |
| | |
| ggsave("Tautitaakka terveysmenojen funktiona maittain.svg", width=9, height=5)
| |
| | |
| pl <- plot_ly(tmp, x=~Y2015, y=~val/100000,type="scatter", mode="markers", hovertext=~Country) %>% #, text=~label,color=~label) %>%
| |
| layout(
| |
| title="Tautitaakka terveysmenojen funktiona maittain 2015",
| |
| xaxis=list(title="Terveydenhuoltomenot per henkilö (USD)"),
| |
| yaxis=list(title="Tautitaakka per henkilö (DALY)")
| |
| )
| |
| | |
| pl
| |
| # pushIndicatorGraph(pl, 103)#, API_KEY= apikey)
| |
| </rcode>
| |
| | |
| == Viitteet ==
| |
| | |
| <references/>
| |
| | |
| * Bruce P. Lanphear, Richard Hornung, Jane Khoury, Kimberly Yolton, Peter Baghurst, David C. Bellinger, Richard L. Canfield, Kim N. Dietrich, Robert Bornschein, Tom Greene, Stephen J. Rothenberg, Herbert L. Needleman, Lourdes Schnaas, Gail Wasserman, Joseph Graziano, and Russell Roberts. (2005) Low-Level Environmental Lead Exposure and Children’s Intellectual Function: An International Pooled Analysis. Environmental Health Perspectives. 1 July 2005 https://doi.org/10.1289/ehp.7688
| |
| * Johanna Suomi, Pirkko Tuominen, Jukka Ranta, Kirsti Savela. (2015) Riskinarviointi suomalaisten lasten altistumisesta elintarvikkeiden ja talousveden raskasmetalleille. Eviran tutkimuksia 2/2015. [https://www.ruokavirasto.fi/globalassets/tietoa-meista/julkaisut/julkaisusarjat/tutkimukset/riskiraportit/riskinarviointi-suomalaisten-lasten-altistumisesta-elintarvikkeiden-ja-talousveden-raskasmetalleille.pdf]
| |
| * EFSA. Lead dietary exposure in the European population. EFSA Journal 2012;10(7):2831 {{doi|10.2903/j.efsa.2012.2831}}
| |
| | |
| == Katso myös ==
| |
| | |
| * {{#l:Ruori-hankkeen tautitaakka-arviot.pptx}}
| |
| * {{#l:Ruori.zip}} Virallinen Ruori-malliajo, josta on otettu luvut ja kuvat Ruori-loppuraporttiin.
| |